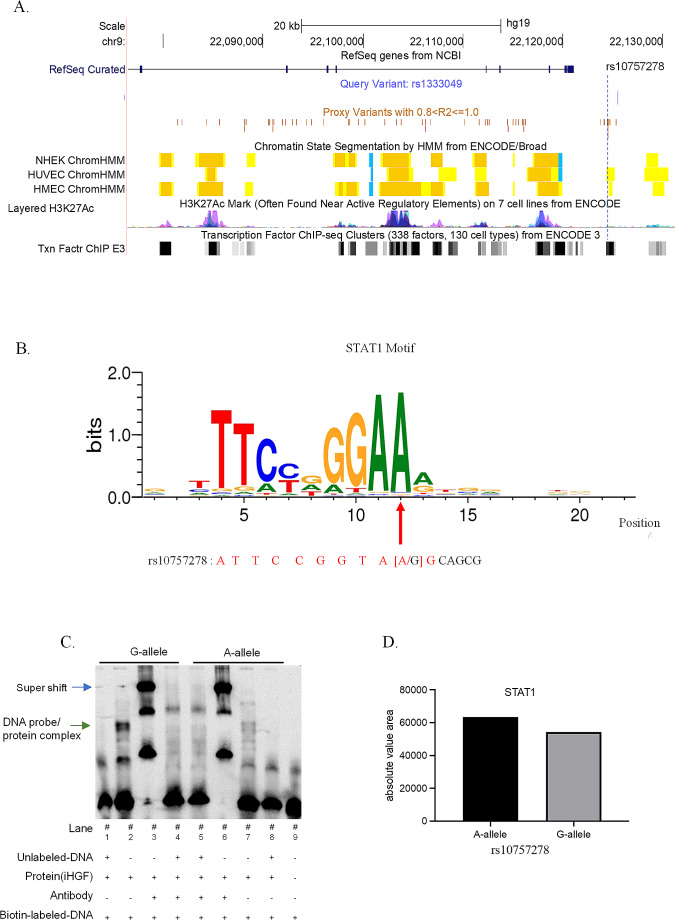
Fig. 2.
rs10757278 is a functional SNP within the chr9p21.3 risk haploblock and localizes to a STAT1 binding site. (A) 55 common SNPs (MAF ≤ 0.05) in CEU and GBR populations in strong LD with the GWAS lead SNP rs1333049 and 24 SNPs located within chromatin elements that correlate with regulatory functions of gene expression, which is indicated by chromatin state segmentation for 3 cell types (data from ENCODE; orange = predicted strong enhancer, yellow = weak enhancer, blue = insulator). Some proxy SNPs locate in H3K4me1 and H3K27ac methylation marks, which are often associated with regulation of gene transcription and within TFBS that were determined from ENCODE ChIP-Seq data. The position of rs10757278 is marked with a dashed line. (B) The DNA sequence at rs10757278-A allele shares a matrix similarity of 95% with the STAT1 transcription factor (TF) binding motif. (C) EMSA was performed with rs10757278 allele-specific oligonucleotide probes and nuclear protein extract from gingival fibroblast cells. Binding of STAT1 antibody to allele-specific probes is shown in lane 2 and 7. The supershift caused by STAT1 antibody binding to the DNA probe-protein complex is seen in lane 3 and 6. Unlabeled DNA was added in lanes 1 and 8 to verify that the band shift was antibody-specific. (D) Absolute value area of the antibody-specific bands. In the background of rs10757278-G allele, STAT1 binding to the allele-specific oligonucleotide probe was reduced 14.4% compared to the A-allele