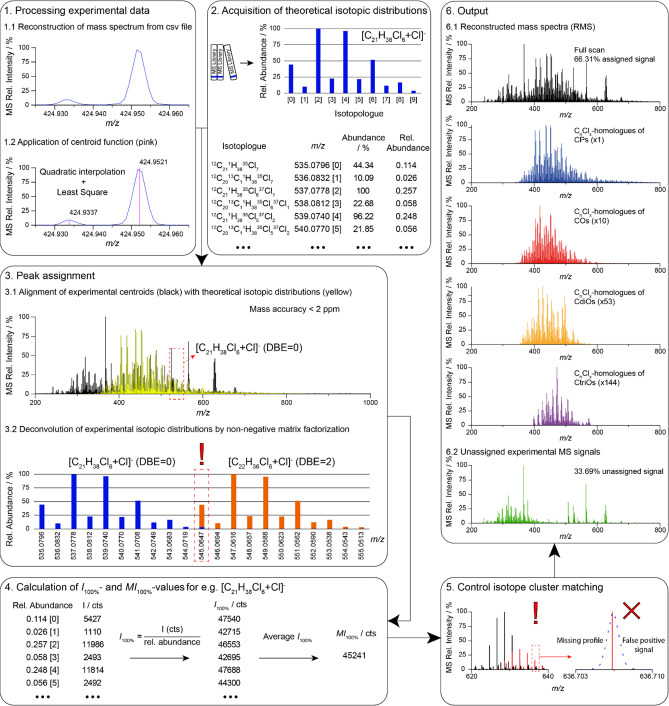
Figure 1.
Workflow of CP-Hunter. Profile-mode MS data are translated into centroids (1). Theoretical isotopic distributions of CPs and their transformation products are obtained from ChemCalc library (2).28 Interfering signals are deconvoluted from one another by non-negative matrix factorization (3).29 Carbon- and chlorine-homologue abundances (MI100%-values) are calculated based on all signals (I100%-values) of the isotope cluster (4). Evaluation of deconvoluted isotope clusters is recommended for low-abundant compounds to identify false positive signals (5). Reconstructed mass spectra (RMS) containing assigned signals to CPs, COs, CdiOs and CtriOs compound classes and not assigned signals are obtained (6).