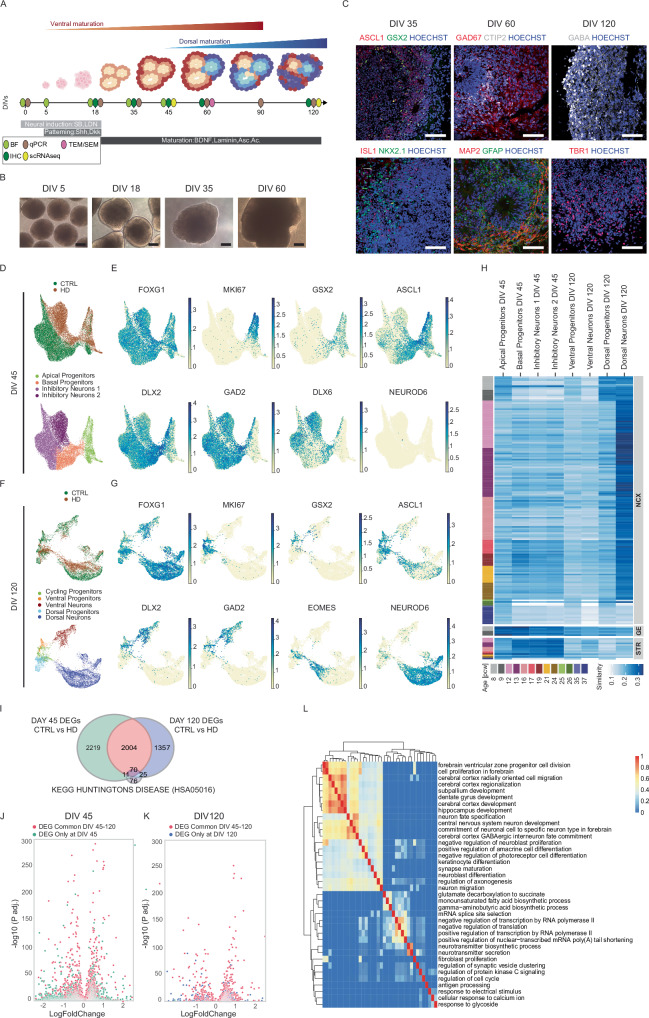
Fig. 1. Telencephalic organoids present ventral and dorsal identities with impaired neuronal transcriptional signatures in HD.
A Scheme of the protocol for telencephalic organoids. Created with BioRender.com. released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Abbreviations: BF=brightfield, IHC=immunohistochemistry, qPCR=quantitative PCR, scRNAseq=single cell RNA sequencing, TEM/SEM=transmission and scanning electron microscopy. B Brightfield images during differentiation. Scale bars = 250 µm C Immunohistochemistry analyses of telencephalic organoids during differentiation. At DIV 35 for ASCL1, GSX2, ISLT1 and NKX2.1; at DIV 60 for GAD67, CTIP2, GFAP, MAP2; at DIV 120 for GABA and TBR1. Scale bars = 100 µm D UMAP plots of scRNAseq analyses on CTRL (20CAG) and HD (56CAG) organoids at DIV 45 showing genotypes and clusters of subpopulations E UMAP plots for specific markers of telencephalic development (FOXG1) with progenitors (MKI67) and ventral identity (GSX2, ASCL1, DLX2, GAD2, DLX6) or dorsal identity (NEUROD6) F UMAP plots of scRNAseq analyses on CTRL (20CAG) and HD (56CAG) organoids at DIV 120 showing the genotypes and clusters of subpopulations G UMAP plots for specific markers of telencephalic development (FOXG1) with progenitors (MKI67) and ventral identity (GSX2, ASCL1, DLX2, GAD2) or dorsal identity (EOMES, NEUROD6) H Voxhunt heatmap of similarity between individual subpopulations of our organoids and human fetal brain by mapping onto BrainSpan human transcriptomic dataset I Venn diagram of DEGs between CTRLs and HD organoids showing which are in common between DIV 45, DIV 120, and KEGG pathway associated to HD (Wilcoxon test, two-sided, p < 0.05) J, K Volcano plot of p-value adjusted (p.adj.) for DEGs at DIV 45 or DIV 120, Wilcoxon test two-sided L Heatmap showing the results of gene ontology analysis of the common DEGs where the enriched GO terms are grouped according to their semantic similarity. (Biological Replicates for scRNAseq: At DIV 45 N=3 individual organoids per genotype and at DIV 120 N=a pool of 10 organoids from 2 cell lines per genotype. See fig. S1).