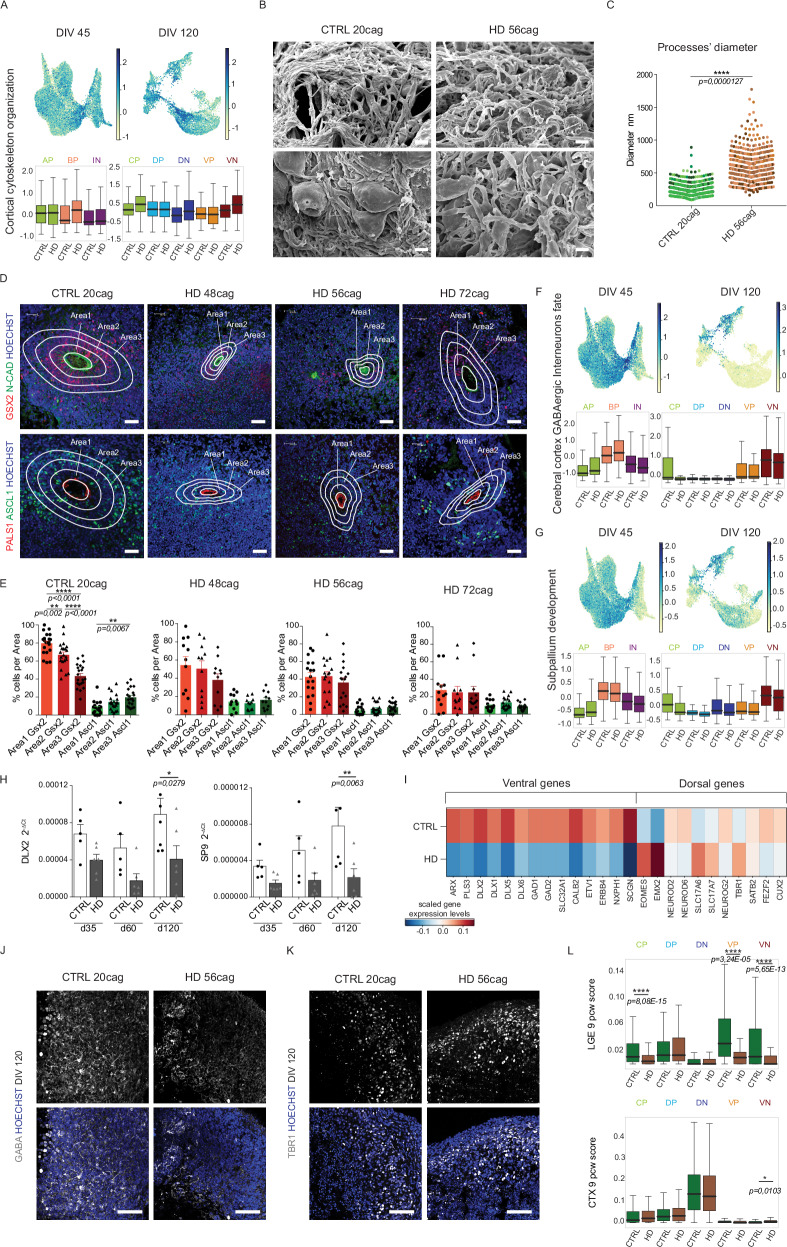
Fig. 2. Self-organization and ventral maturation are altered in HD organoids.
A UMAP and boxplot of score of DEGs of term “cortical cytoskeleton organization” upregulated in HD organoids at both time points. Abbreviations: AP=Apical Progenitors; BP=Basal Progenitors; IN=Inhibitory Neurons; CP=Cycling Progenitors; DP=Dorsal Progenitors; DN=Dorsal Neurons; VP=Ventral Progenitors; VN=Ventral Neurons B Scanning electron microscopy (SEM) images of CTRL and HD organoids at DIV 60. Scale bars = 5 µm C Diameter of neuronal processes (Dots shown are all the multiple processes measured in N = 6 organoids of independent biological replicates, but statistics is performed on the mean of processes per organoid. Dots coloured based on the 6 replicates). Unpaired two-tailed t-test, ****p<0,0001) D Immunohistochemistry analyses of CTRL (parental and 20CAG) and HD (48, 56 and 72 CAG) organoids at DIV 45-60 for GSX2, ASCL1, N-CADHERIN and PALS1. Scale bar 30 µm E Numbers of GSX2+ or ASCL1+ cells in each area (Manual counting. Anova One Way, Bonferroni post test, **p < 0,01, ****p < 0,0001. N ≥ 11 VZ-like structures, derived from 10 organoids of 2/3 biological replicates) F, G UMAP and boxplot of score of DEGs of terms “cerebral cortex GABAergic interneurons” and “subpallium development” downregulated in HD organoids H q-PCR for DLX2 and SP9 on CTRL and HD organoids at DIV 35, 60, and 120. (N = 5 independent biological replicates where each one is a pool of 4 organoids; error bars represent ± SEM; Anova One Way, Bonferroni post test, *p < 0,05; **p < 0,01) I Heatmap of gene expression of key dorso-ventral telencephalic markers in CTRL and HD organoids at DIV 120 J, K Immunohistochemistry analyses of CTRL and HD organoids at DIV 120 for GABA and TBR1. Scale bar 100 μm L Boxplots of the signature score of each subpopulation of organoids at DIV 120 and the bulk RNAseq from human fetal brain LGE and CTX (cortex) at 9pcw39 (Wilcoxon test, two-sided, Bonferroni post test, ****p<0,0001). For A, F, G, and L the box plots show the median (centre line), upper and lower quartiles (box limits), and the highest and lowest values within 1.5× the IQR of the nearest hinge (whiskers). Source data are provided as a Source Data file.