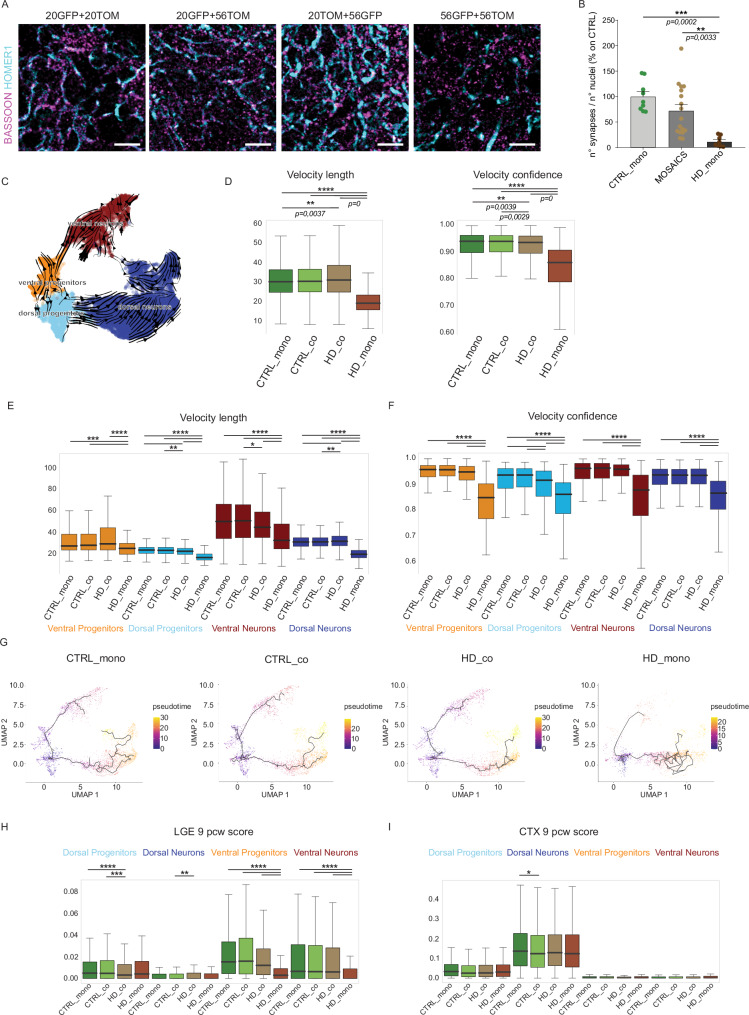
Fig. 4. HD cells recover maturation and fate determination when grown with CTRL cells.
A Immunohistochemistry analyses of CTRL_mono (20GFP+20TOM) co-culture mosaic organoids (20GFP+56TOM and 20TOM+56GFP) and HD_mono (56GFP+56TOM) organoids at DIV 120 of differentiation for BASSOON and HOMER1. Scale bar 20 µm B Automatic quantification of numbers of synapses as BASSOON/HOMER1 co-localizing puncta in each condition. As it was not possible to quantify single GFP or TOM cells overlapping with the puncta staining, the quantification reports the total number of synapses in the co-culture mosaic condition (N = 8 organoids from 2 independent biological replicates. Data are normalized over the mean of CTRL values, error bars represent ± SEM. Anova One Way, Bonferroni post test. ***p < 0.001, **p < 0.01). C Differentiation trajectories of all mosaic organoids together, inferred with the method of RNA velocity D Length and confidence of the velocity vectors in each culture sample (Wilcoxon test, two-sided, Bonferroni post test, **p < 0,01; ****p < 0,0001) E, F Length and confidence of the velocity vectors in each individual subpopulation (Wilcoxon test, two-sided, Bonferroni post test, *p < 0,05; **p < 0,01; ***p < 0,001; ****p < 0,0001) G Pseudotime analysis in each culture sample performed using Monocle3 H, I Boxplots for the signature score (as described in methods) of comparison between our mosaic organoids over the bulk RNAseq from human fetal LGE and CTX (cortex) at 9pcw39, focusing on individual subpopulation of each sample (Wilcoxon test, two-sided, Bonferroni post test, *p < 0,05; **p < 0,01; ***p < 0,001; ****p < 0,0001). For D, E, F, H, and I the box plots show the median (centre line), upper and lower quartiles (box limits), and the highest and lowest values within 1.5× the IQR of the nearest hinge (whiskers). For E, F and H, I exact p-values can be found in Supplementary Table 2 inside the Supplementary Information. Source data are provided as a Source Data file.