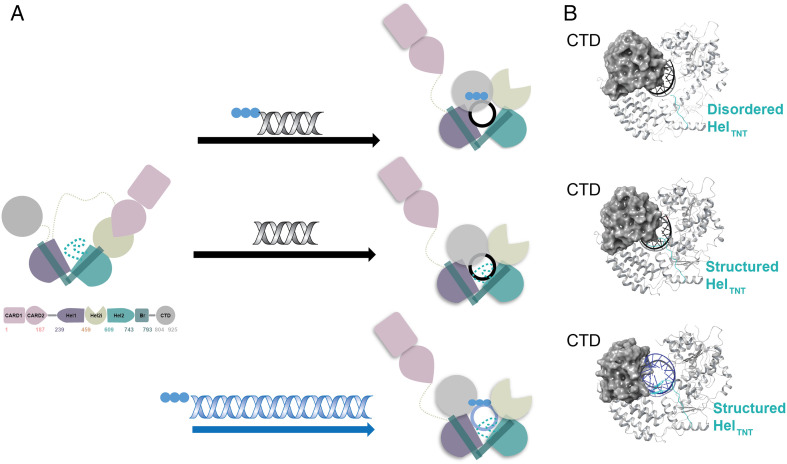
Figure 1.
Schematic representation of structural rearrangements of RIG-I upon binding distinct types of RNA. (a) Conformational changes and domain rearrangements that RIG-I can undergo when binding RNA. All RNA binding leads to the ejection of the N-terminal CARDs, independent of nucleotide presence. Top panel: Binding of short 5′-triphosphate dsRNA leads to rearrangement of the helicase core, an unstructured HELTNT loop (sequence N664-RGKTNQNTG-C672), and a CTD orientation that caps the 5′-end of RNA. Middle panel: Binding of short dsRNA ending with a 5′-hydroxy group leads to rearrangement of the helicase core, a structured HELTNT loop, and a CTD orientation that caps the 5′-end of RNA. Bottom panel: Binding of long dsRNA (here 112 bp) leads to rearrangement of the helicase core, a structured HELTNT loop, and the CTD rotated away from the central axis of RNA as in the case of MDA5. (b) Cartoon representation of differences seen for the CTD and HELTNT loop conformations when binding the respective types of RNA. The CTD is represented as a gray sphere, and short versus long RNA are color-coded black and blue, respectively. The apparent displacement of the CTD upon binding long RNA can be observed (bottom panel). The HELTNT loop is indicated in teal, and it is disordered when short 5′-triphosphate is bound to RIG-I (top panel); in the presence of short RNA ending with 5′-hydroxy group (middle panel) or long RNA (bottom panel), the loop becomes structured. CARDs: caspase activation and recruitment domains; CTD: C-terminal domain; dsRNA: double-stranded RNA; MDA5: melanoma differentiation-associated gene 5; RIG-I: retinoic acid-inducible gene I.