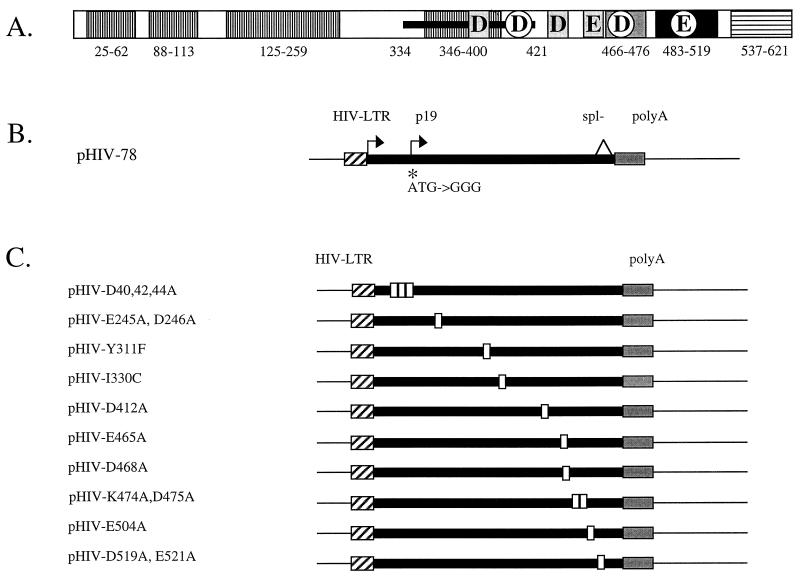
FIG. 1.
AAV Rep78. (A) Schematic diagram of previously described functional domains (see text) within Rep78/68. Regions described as required for binding (vertical stripes), nuclear localization (black box) (amino acids 483 to 519), oligomerization (gray box) (amino acids 466 to 476), and ATPase and helicase activities (black line) (amino acids 334 to 421) and the unique region between Rep78 and Rep68 (horizontal stripes) (amino acids 537 to 621) are indicated. Overlapping DD35E motifs are indicated as boxed residues corresponding to one motif at D368, D429, and E465 and as circled residues corresponding to a second motif at D412, D468, and E504. (B) Eucaryotic expression cassette (pHIV-78) for generating Rep78 proteins. Protein expression is under control of the HIV LTR promoter and the poly(A) signal from simian virus 40. The initiation codon (ATG) at p19 was changed to GGG to eliminate expression of Rep52 and Rep40, and the splice donor site was modified (spl−) as described previously (74) to eliminate expression of Rep68. (C) Positions of point and clustered alanine mutations (open boxes) within the pHIV-78 expression plasmid. Amino acid changes are indicated by the nomenclature for each construct.