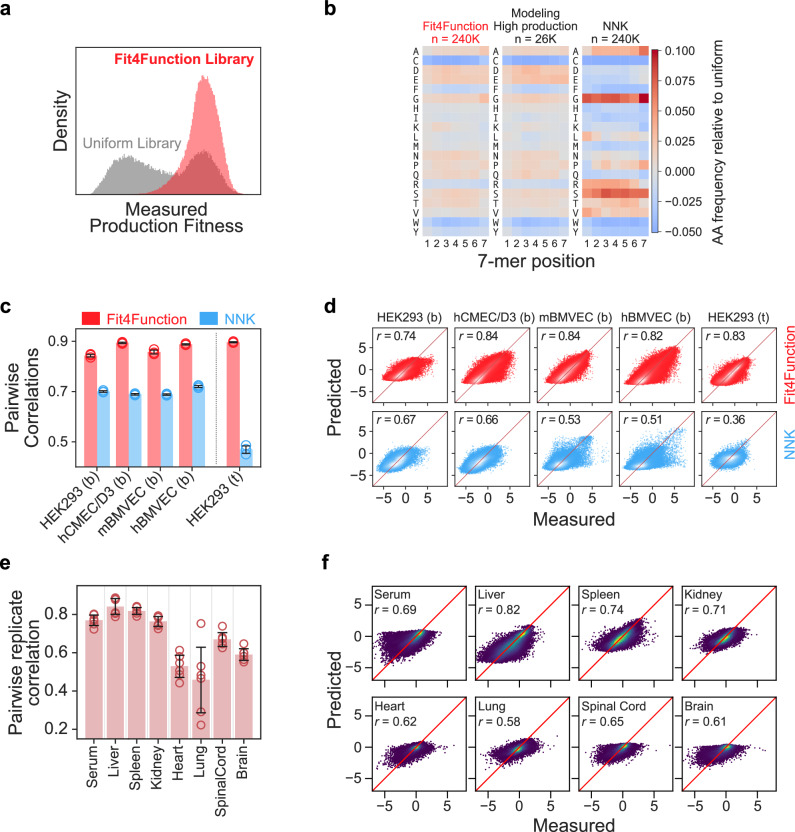
Fig. 3. Fit4Function libraries uniformly sample the production-fit space and enable more accurate functional screening and prediction.
a The distributions of the measured fitness scores are shown for 100K randomly sampled variants from the Fit4Function library versus the uniform modeling library. b The amino acid distribution by position for the variants in the Fit4Function library, production-fit distribution of the modeling library, and 240K most abundant sequences in an NNK library are shown. c The pairwise Pearson correlations among biological triplicates across functional screens (mean ± s.d.; one-tailed paired t-test, n = 5 screens; p = 0.0065) using the Fit4Function library (240K) versus an NNK library (top 240K variants) are shown. hCMEC/D3: human brain endothelial cell line, mBMVEC: primary mouse brain microvascular endothelial cells, hBMVEC: primary human brain microvascular endothelial cells, HEK293: HEK293T/17 cells. Binding and transduction are indicated by ‘b’ and ‘t’, respectively. d The measured versus predicted functional fitness (log2 enrichment scores) for models trained on Fit4Function versus NNK library data are shown. e The replication quality (mean ± s.d.) between pairs of animals (n = 6 pairs across four animals) for the Fit4Function library biodistribution in eight tissues is shown. f The prediction performance of models trained on the in vivo biodistribution of the Fit4Function library across eight organs is shown. Source data are provided as a Source Data file.