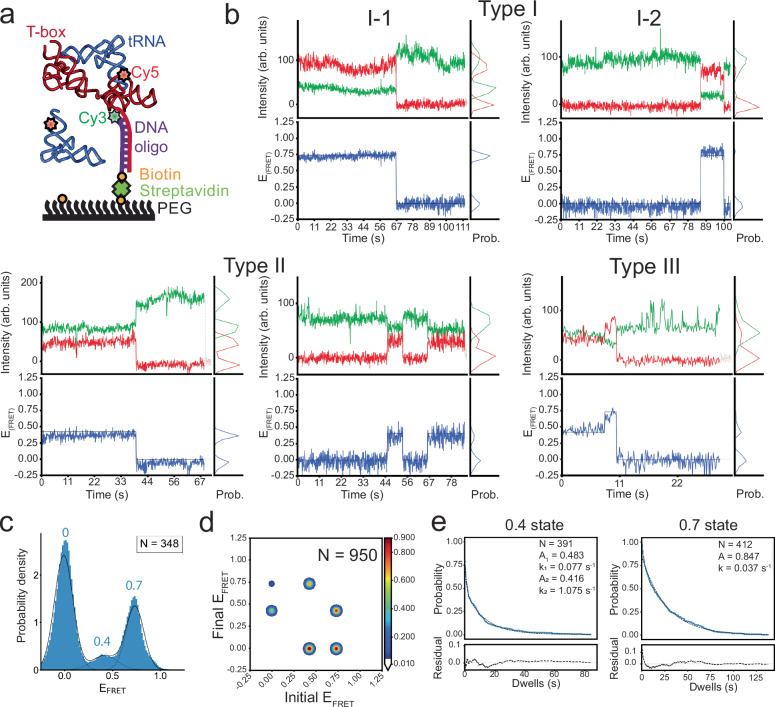
Fig. 2. smFRET experiments show binding of tRNAIle to the wild-type Mtb-ileS T-box riboswitch.
a Schematic diagram illustrating the smFRET experiment. The T-box was anchored to the surface via hybridization to a DNA oligonucleotide (purple) biotinylated on the 5’ end and Cy3-labeled at the 3’ end. The tRNAIle was labeled on the 5’ end with Cy5. Cartoon based on a similar cartoon from19. b Representative Cy3 (green) and Cy5 (red) fluorescence intensity (top) and smFRET trajectories (bottom) of the Mtb-ileS T-box riboswitch/tRNAIle. The smFRET efficiency was calculated as (ICy5/(ICy3-ICy5)). Three types of trajectories were observed: Type I trajectories only show FRET efficiency around 0.7, Type II trajectories only show FRET values around 0.4, and Type III trajectories sample the 0.4 and 0.7 states. c FRET efficiency histogram of the data. Dotted black lines show the individual populations obtained from the consensus HMM modeling, while the model’s population-weighted set of efficiency distributions is plotted with a solid black line. N reports the number of traces included in the histogram. d Transition density plot constructed using the idealized Viterbi paths modelled to the entire dataset using tMAVEN26. The contour level colors report the normalized counts. N reports the total number of transitions. e Dwell time survival plot for the 0.4 and 0.7 FRET states. The data were fitted using either a single (0.7 state) or double (0.4 state) exponential decay function. The number of events (N) used for the analysis and the fitting parameters are shown. Data are shown in blue and the fitting curves are shown in dotted black lines. The fit residuals are shown in the bottom plot. Detailed fitting results are reported in Supplementary Table VI. Source data are provided as a Source Data file.