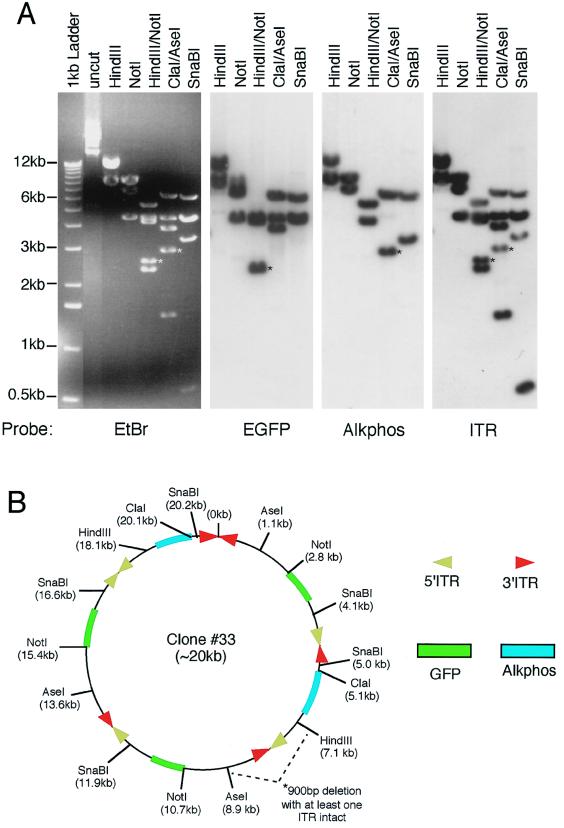
FIG. 5.
Structural analysis of bifunctional concatamer circular intermediates. To fully characterize the nature of GFP- and Alkphos-coexpressing circular intermediates, detailed structural analyses were performed by using restriction enzyme mapping and Southern blot hybridization against GFP, Alkphos, and ITR 32P-labeled probes. (A and C) Results from Southern blot analysis of plasmid clones 33 (A) and 5 (C) are given as representative examples of circular intermediates isolated from 80- and 35-day Hirt DNAs of rAAV-infected muscle, respectively. Agarose gels were run in triplicate for each of these clones, and Southern blot filters were hybridized with one of the three DNA probes as indicated below each autoradiogram. Molecular sizes are indicated to the left of the ethidium bromide (EtBr)-stained agarose gel, and restriction enzymes are marked on the top of each gel or filter. (B and D) Deduced structures of plasmid clones 33 and 5, respectively, based on Southern blot analysis. For ease of comparison with the restriction maps of the viral genomes given in Fig. 1A, the positions of restriction enzyme sites are marked with the indicated orientation of intact viral genomes. However, in clone 33 a deletion occurred between the AseI and HindIII site of a head-to-tail array between AV.Alkphos and AV.GFP3ori, as reflected by a 900-bp reduction in the anticipated sizes of HindIII/NotI and ClaI/AseI fragments (marked by asterisks in panel A). Furthermore, the SphI site flanking an ITR was ablated in clone 5 (bands affected by this deletion are marked by asterisks in panel C). These deletions are not reflected in the size markings of the overall concatamer, since the exact region involved and/or the size of the deletion is unclear. Additionally, chemical sequence evidence for rescued circular intermediates suggests that the predominant form of ITR arrays may be in a double-D structure (i.e., one ITR flanked by two D sequences rather than two ITRs) (unpublished data), and hence ITR arrays containing fragments may appear 147 bp shorter than indicated. However, to more easily depict the orientations of viral genomes, we have indicated the positions of 5′ and 3′ ITRs rather than representing a single ITR at these junctions.