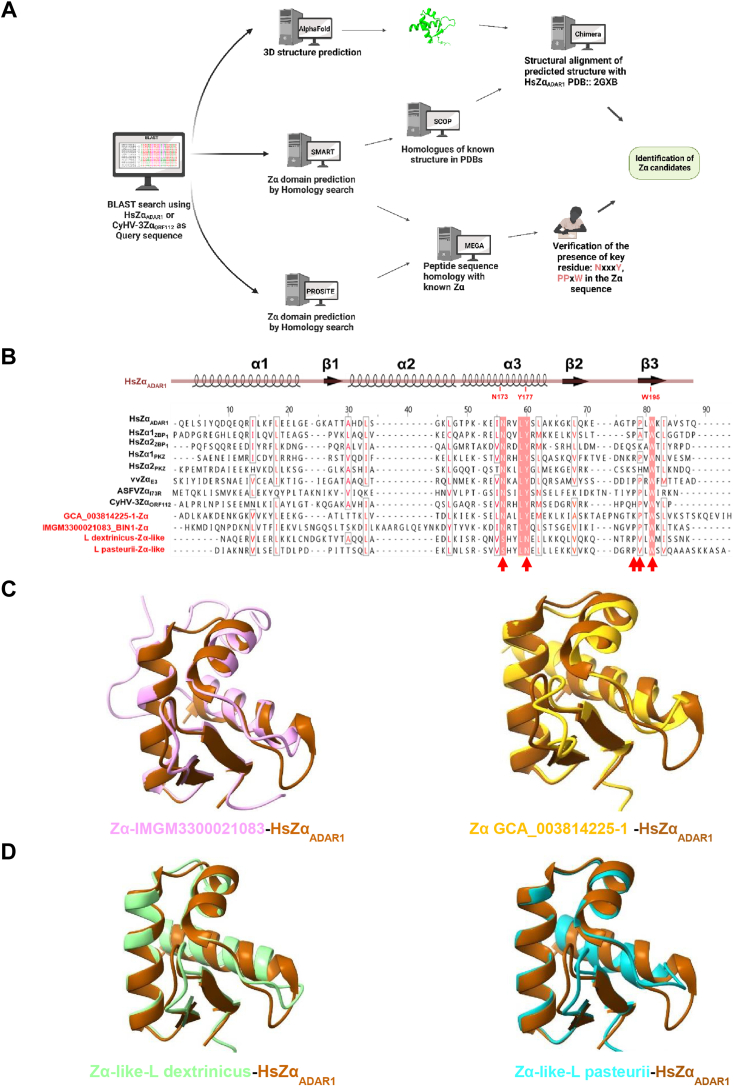
Figure 1.
Prediction of new Zα candidates. A, the summary of the workflow for identifying Zα candidates. B, alignment of two predicted Zα and two Zα-like with known Zα domains from human and mouse ADAR1 (HsZαADAR1), human ZBP1 (HsZαZBP1), and viral from vaccinia virus and cyprinid-herpesvirus-3 (vvZαE3 ASFV ZαI73R CyHV-3Zα112). The consensus structure Zα domain is shown at the top. The key DNA-interacting residues in ADAR1 (corresponding to Asn174, Tyr177, P192, P193, and Trp195 in HsZαADAR1) are pointed with red arrows. Hydrophobic residues in the hydrophobic core are highlighted in red front. C, structural alignment of AlphaFold predicted structure of Zα candidates with HsZαADAR1 (brown, PDB ID 2GXB). D, structural alignment of AlphaFold predicted structure of Zα-like candidate with HsZαADAR1 (brown, PDB ID 2GXB).