Fig. 1|. The genes involved in distal cholesterol synthesis differentially regulate ferroptosis.
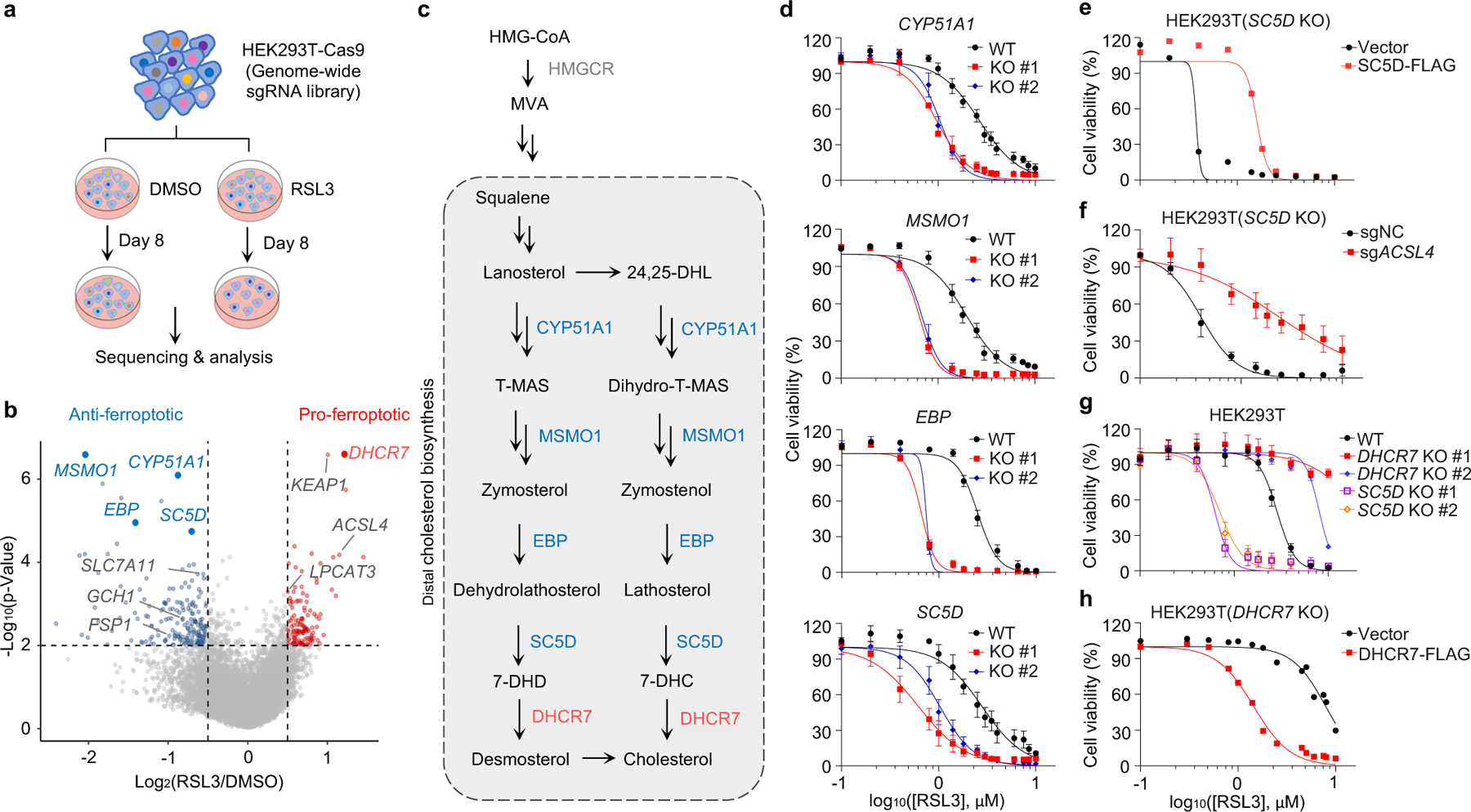
a, CRISPR screening workflow for identification of ferroptosis regulators. b, Volcano plot of top-ranked genes identified in RSL3-treated HEK293T-Cas9 cells. Genes that meet the cutoff criteria (P<0.01 and LFC>0.5 or <-0.5) are indicated as follows: genes (grey) represent known ferroptosis regulators and genes (blue and red) are involved in cholesterol biosynthesis. The p-Value and LFC are generated by the MAGeCK-test module using modified robust ranking aggregation (α-RRA) analysis. Top screen hits are shown in red (pro-ferroptotic) or blue (anti-ferroptotic). c, Schema of distal cholesterol biosynthesis pathway. d, Cell viability of HEK293T cells expressing sgRNAs targeting negative control (WT), CYP51A1 (CYP51A1 KO), MSMO1 (MSMO1 KO), EBP (EBP KO), or SC5D (SC5D KO), after treatment of indicated concentrations of RSL3 for 6–8 h. e, Cell viability of SC5D KO HEK293T cells expressing vector or SC5D cDNA after treatment of RSL3 for 6–8 h. f, Cell viability of SC5D KO HEK293T cells expressing sgRNAs targeting negative control or ACSL4 after treatment of RSL3 for 6–8 h. g, Cell viability of HEK293T cells expressing sgRNAs targeting negative control, DHCR7 (DHCR7 KO), or SC5D (SC5D KO) after treatment of RSL3 for 6–8 h. h, Cell viability of DHCR7 KO HEK293T cells expressing vector or DHCR7 cDNA after treatment of RSL3 for 10–12 h. Data are mean ± s.d. of n=3 biological replicates (d, f, g). For d-h, n=3 biological replicates (d, f, g) and n=2 biological replicates (e, h) are representative of three independent experiments.
