Extended Data Fig. 2|. The distal cholesterol biosynthesis genes regulates ferroptosis.
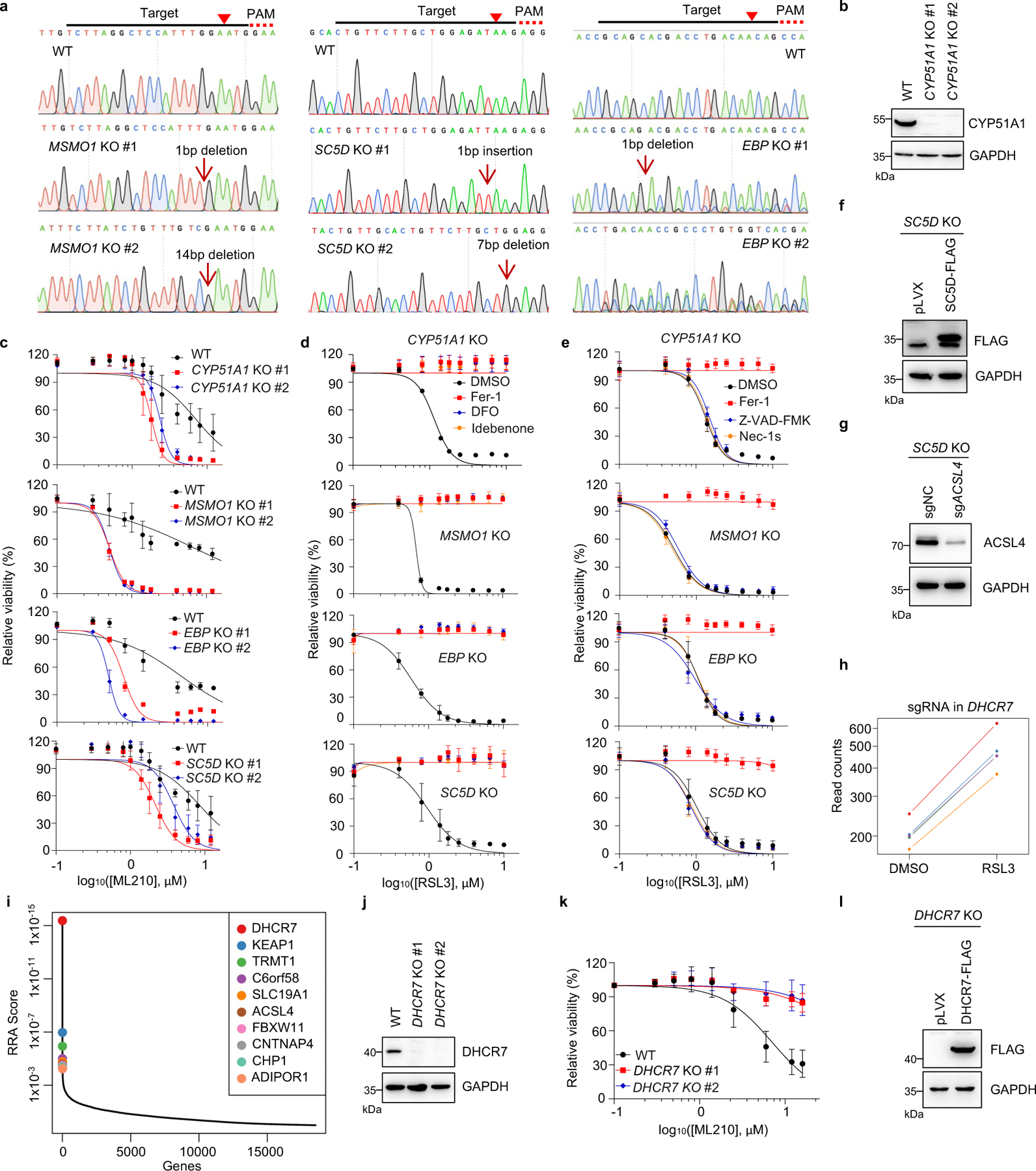
a, Genotyping of subclones generated from HEK293T cells expressing sgRNAs targeting MSMO1, SC5D and EBP by Sanger sequencing. b, The protein level of CYP51A1 in CYP51A1 KO HEK293T cells was analyzed by Immunoblotting. c, Cell viability of WT and CYP51A1, MSMO1, EBP, SC5D KO HEK293T cells treated with ML210 for 8–10 h. d, e, Cell viability of WT and CYP51A1, MSMO1, EBP, SC5D KO HEK293T cells treated with RSL3 for 6–8 h following pretreatment of DMSO, Fer-1 (1 μM), DFO (20 μM) and Idebenone (10 μM) (d) or Fer-1 (1 μM), Z-VAD-FMK (20 μM) and Nec-1s (20 μM) (e). f, The protein level of SC5D-FLAG in SC5D KO HEK293T cells expressing vector and SC5D-FLAG was analyzed by Immunoblotting. g, The protein level of ACSL4 in SC5D KO HEK293T cells expressing sgRNAs targeting ACSL4 was analyzed by Immunoblotting. h, The normalized read counts of sgRNAs by DHCR7, generated by the MAGeCK-test module. n=5 sgRNAs. i, Integrative analysis of pooled CRISPR genetic screening using MAGeCK and identification of top candidate genes that were assigned with values generated from positive selection (pro-ferroptotic) by modified robust ranking aggregation (α-RRA) analysis. j, The protein level of DHCR7 in WT and DHCR7 KO HEK293T cells was detected by Immunoblotting. k, Cell viability of WT and DHCR7 KO HEK293T cells treated with ML210 for 8–10 h. l, The protein level of DHCR7-FLAG in DHCR7 KO HEK293T cells expressing vector and DHCR7-FLAG analyzed by Immunoblotting. For b-g, j-l, data are representative of three independent experiments. Data are mean ± s.d. of n=3 biological replicates.
