Figure 4: Generalist and specialist metabolism differs in expected and unexpected ways.
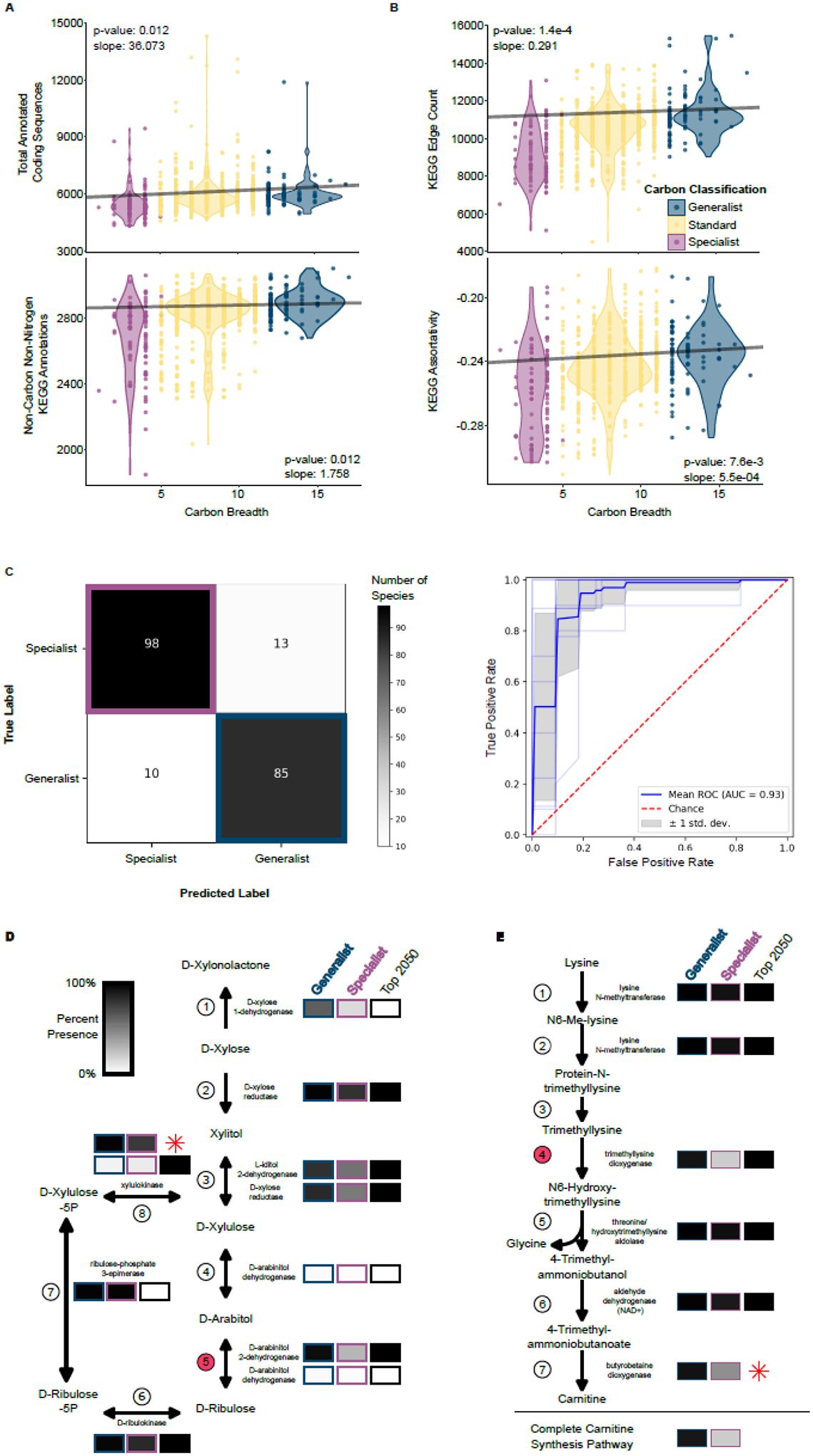
A. Total annotated coding sequences (top) and total number of annotated KEGG ortholog groups (KOs; bottom) are both positively and significantly correlated with carbon niche breadth using a Phylogenetic Generalized Least Squares (PGLS) analysis. One outlier with a predicted number of coding sequences is not visualized but was included in the analysis (Magnusiomyces magnusii, number of protein-coding genes = 20,704, carbon niche breadth = 9).
B. Two KEGG network statistics were significantly and positively correlated with carbon niche breadth when taking into account phylogenetic relatedness (PGLS). KEGG Edge Count (top) and KEGG Assortativity (bottom) were both elevated in carbon generalists.
C. Yeasts were classified into generalists and specialists using a machine learning algorithm trained on the KOs. The correct classification occurred in 88% of specialists and 89% of generalists. The ROC analysis suggests that both the sensitivity and specificity of our model is excellent (AUC=0.93).
D. Multiple reactions in the pentose and glucuronate interconversions pathway were important in classifying yeasts into generalists and specialists as determined by the leave-out analysis, which identified 2,050 informative KOs (black boxes.) Boxes are shaded as the percent of each carbon classification with at least one enzyme in that step of the reaction. The reaction with the third highest relative importance in the machine learning analysis is shown in Step 5 and is facilitated by D-arabinitol 2-dehydrogenase. Interestingly, experimental studies suggest that yeast D-arabinitol 2-dehydrogenase is also capable of completing the reaction in Step 4 (93). Step 8 was among the top features used in the machine learning analysis, despite the fact that KEGG only partially annotated this gene. The xylulokinase encoded by yeast XYL3 is well studied (58). Therefore, we re-annotated the XYL3 gene and have shown its relative abundance (red star).
E. The carnitine biosynthesis pathway includes multiple reactions that are important for classifying carbon generalists and specialists. The reaction in Step 4 had the fourth highest relative importance in the machine learning classification of carbon classification. Step 7 was not annotated by KEGG in any of our yeasts, but this step had been previously characterized in Candida albicans as being facilitated by the trimethyllysine dioxygenase enzyme encoded by BBH2 (64). We re-annotated BBH2 using this reference sequence and calculated the relative abundance in each carbon classification (red star). Finally, we determined the number of yeasts that could hypothetically complete the lysine to carnitine biosynthesis pathway.
