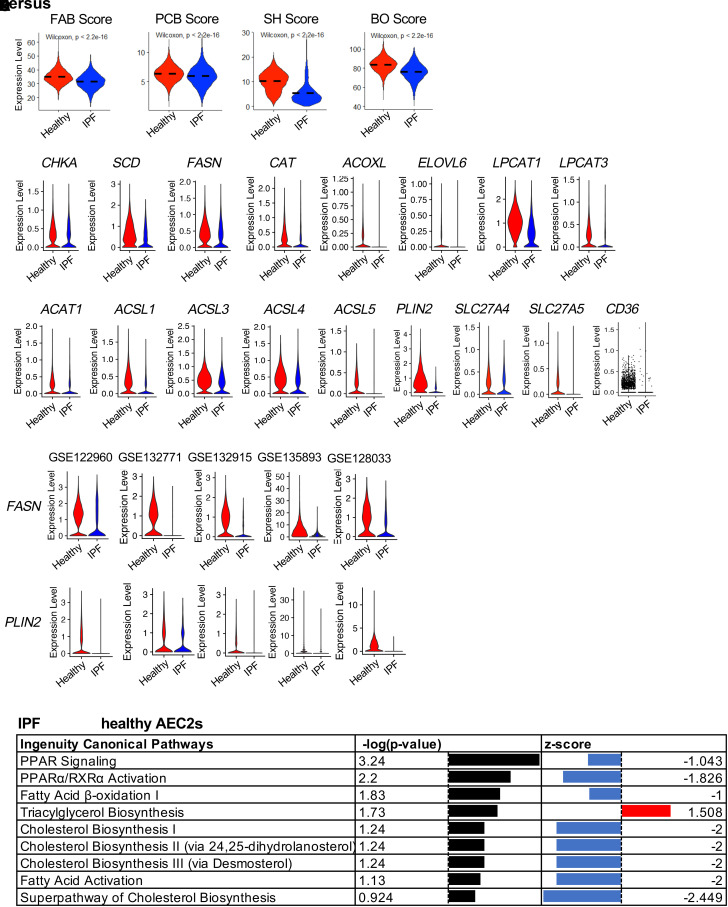
Figure 1.
Dysregulated lipid metabolism of idiopathic pulmonary fibrosis (IPF) alveolar type II cells (AEC2s). (A) Activation scores of fatty acid biosynthesis (FAB), phosphatidylcholine biosynthesis (PCB), β-oxidation (BO), and surfactant homeostasis (SH) of AEC2s from healthy and IPF lungs (red, healthy; blue, IPF) analyzed from dataset GSE157996. (B and C) Violin plots of expression of lipid metabolism–related genes (B) and fatty acid β-oxidation, lipid droplet, and lipid uptake–related genes (C) in healthy and IPF AEC2s (red, healthy; blue, IPF). (D) Violin plots of expression of FASN and PLIN2 (perilipin 2) with published single-cell RNA sequencing (scRNA-seq) datasets. (E) IPA pathway analysis of human AEC2s from IPF versus healthy lungs analyzed with dataset GSE157996. PPARγ = peroxisome proliferator activated receptor γ. IPA = ingenuity pathway analysis.