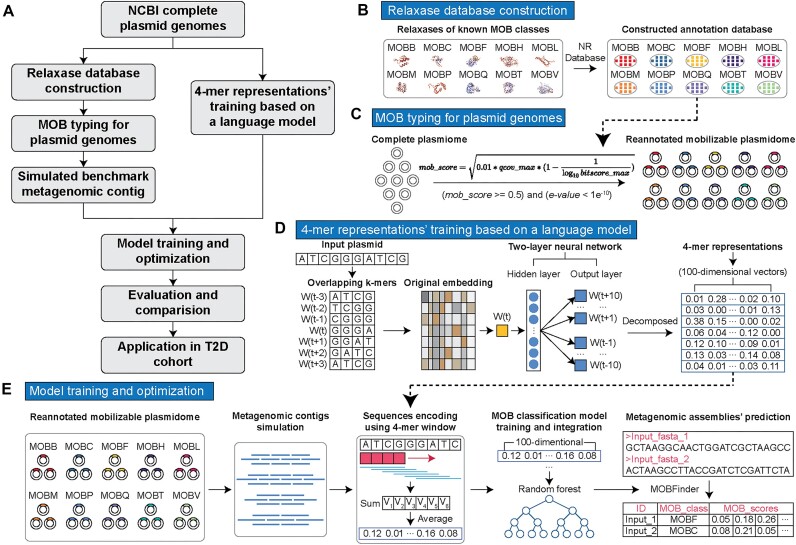
Figure 1:
Flowchart of the technical approach utilized in this study. (A) General workflow of the development and testing of MOBFinder. (B) Using plasmid relaxases with known MOB types as reference sequences, we developed a database of relaxases from the nonredundant (NR) database representing different MOB types. (C) Utilizing the relaxase database, complete plasmid genomes from the NCBI were subjected to MOB typing. (D) Those complete genomes were also used to train a 4-mer language model using the skip-gram algorithm, allowing each 4-mer to be represented by a 100-dimensional word vector. For a DNA fragment, the average word vector of all 4-mers on its sequence serves as the feature vector for that DNA. (E) We constructed simulated metagenomic contigs from the complete genomes that had been MOB typed as a benchmark and encoded these contigs into word vectors. Then these word vectors were used to train a random forest algorithm. Then the trained model, with metagenomic DNA fragments as input, was used to predict the MOB typing of the corresponding DNA fragment based on its word vectors.