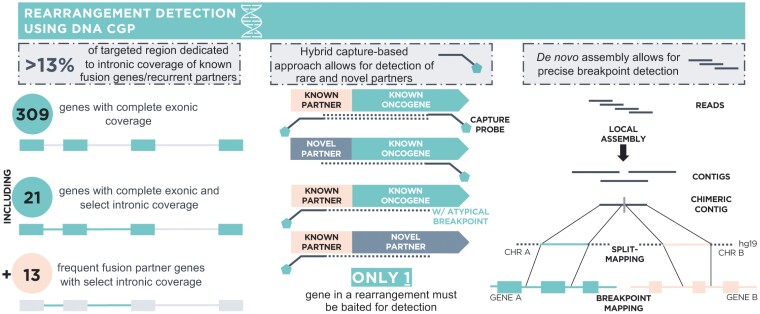
Figure 1.
F1CDx assay design enables robust DNA-based rearrangement detection. Design features of F1CDx that support rearrangement detection include (1) select intronic coverage (Supplementary Table S1) for N = 34 genes including both common fusion genes (N = 21) and frequent fusion partners (N = 13), (2) the use of a hybrid capture-based sequencing approach which only requires that one gene involved in a fusion be baited allowing for detection of known as well as rare and novel fusion partners, and (3) the use of a de novo assembly approach in the bioinformatics pipeline which facilitates precise breakpoint detection compared to reference-based methods.