Abstract
We had previously described six distinct alleles of the glycoprotein B (gB) gene of human herpesvirus 7 (HHV-7). The genetic changes corresponding to these alleles did not affect gB gene transcription or translation in in vitro assays. The study of distinct HHV-7-positive human samples showed preferential associations of some gB alleles with some alleles of two other genes, distantly located on the HHV-7 genome, coding for the phosphoprotein p100 (p100) and the major capsid protein (MCP). Two allele combinations, corresponding to 44 and 31% of the samples studied, respectively, were interpreted as the genetic signatures of two major prototype HHV-7 variants.
Human herpesvirus 7 (HHV-7) was originally isolated from the stimulated CD4+ T cells of a healthy individual (9) and was subsequently characterized as a ubiquitous virus, infecting most human beings (5, 10, 17, 25, 27). This virus was classified in the Betaherpesvirinae subfamily on the basis of its genetic organization (3, 6, 15, 16). Infected saliva is generally considered to be the main source of human transmission (4, 26). Although many diseases have been hypothetically related to HHV-7 infection (1–3, 7, 12, 13, 18, 19, 21, 23, 24), convincing proof and precise knowledge of its pathogenicity are still missing.
The entire genome of HHV-7 has been recently sequenced for two reference strains: JI (16) and RK (15). Comparison of these two strains has shown the high degree of conservation of the HHV-7 genome. However, a previous study had found a restricted polymorphism of the glycoprotein B (gB) gene: five critical positions were identified as the sites of point nucleotide substitutions, and the stable combination of specific changes at these positions allowed us to define six alleles of the gene (8). The distribution of gB alleles varied according to the geographical origin of the samples, suggesting the possibility of using these alleles as indirect markers for the study of population genetics. The reasons why the different gB alleles have emerged and have been maintained in human populations were not clear. The protein gB plays an important role in the early events of virus-cell interaction (11, 20), but the genetic differences between gB alleles were silent at the protein level and did not favor the concept of selection pressure based on distinct phenotypic properties. However, subtle modifications of replication properties due to conformational differences or preferential nucleotide usage could not be ruled out. An alternative and more likely hypothesis was that gB alleles were tightly associated with specific alleles of other genes, these preferential associations being stably transmitted through human generations.
We then decided to explore these two possibilities through recombinant gB expression assays and novel genetic analyses of different HHV-7-positive human samples. The preliminary results shown here confirmed the high conservation of the HHV-7 genome with a limited apparent impact of allele-specific changes on phenotypes. However, these data allowed us to move from the concept of gene alleles to that of HHV-7 variants.
Transcription and translation efficiency of HHV-7 gB alleles.
In order to investigate whether the six gB alleles exhibited a different capacity to be transcribed, in vitro transcription was studied after each allele had been cloned in the plasmid pcDNA3.1 under the control of the T7 promoter. The RNA transcripts, synthesized by means of T7 RNA polymerase with the RiboMAX in vitro transcription kit (Promega, Madison, Wis.) as reported previously (14), had an apparent molecular length of 2,400 bp, as expected (20). Their concentration was estimated by spectrophotometry, and comparison of these concentrations showed no differences between the six alleles (data not shown). Another question was the possibility of differences in the translation of transcripts. To explore that point, a 3.8-kbp chimeric gene consisting of a 2.4-kbp gB gene fused with a 1.4-kbp luciferase gene (luc) (22), was constructed for each of the six alleles and subcloned into the mammalian expression vector pcDNA. Optimal conditions for expression had been previously established by introducing the six-histidine-containing sequence ATGCCGCGGGGTTCT(CAT)6GGTATGGCTAGC upstream of the luciferase gene and studying the expression of (HIS)6-luciferase in plasmid-transfected CHO cells by means of (HIS)6-sequence-specific immunofluorescence assay. CHO cells were transfected with the SUPERFECT transfection reagent (Qiagen, San Diego, Calif.) with plasmids containing the chimeric gB-luc genes, and luciferase activity was determined by using the luciferase assay reagent (Promega). Luciferase activity was similar for the six different chimeric gB-luc genes, suggesting that no difference in translation efficiency was detectable between the six gB gene alleles (data not shown). These results showed that the allele-specific alterations of the gB gene which did not induce any change in the predicted amino acid sequence had no apparent effect on either the transcription or translation of this gene. This result reinforced the general conclusion that the gB gene of HHV-7 was highly conserved and led us to hypothesize that stable gB alleles were related to a more general polymorphism of the HHV-7 genome rather than to specific properties of gB.
Polymorphism of the p100, MCP, and gL genes.
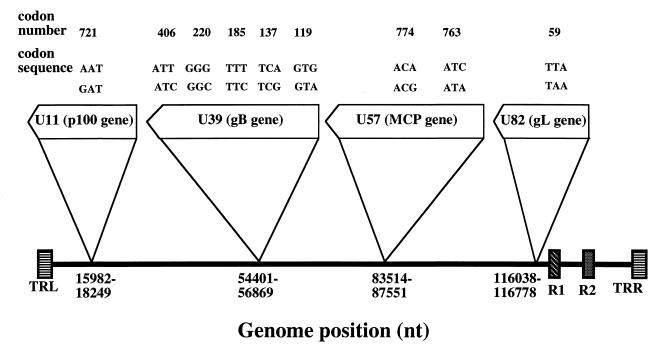
Our study was subsequently focused on three other genes located on distant parts of the HHV-7 genome: the gene coding for the structural phosphoprotein p100, known as open reading frame (ORF) U11 and located at position 15982 to 18249; the gene coding for the major capsid protein (MCP), known as ORF U57 and located at position 83514 to 87551; and the gene coding for glycoprotein L (gL), known as ORF U82 and located at position 116038 to 116778 (Fig. 1). Comparison of the JI (16) and RK (15) sequences revealed relevant differences between these three genes. In the p100 gene, an AAT-to-GAT substitution induced the presence of an MboII restriction site in RK, this site being absent in the case of JI, and the predicted change of an asparagine residue (for JI) into an aspartic acid residue (for RK). In the MCP gene, the codons ATC and ACA (JI) at positions 763 and 774 were changed into ATA and ACG, respectively (RK). Consequently, the corresponding MboII and MunI restriction sites present in JI were absent and had been replaced by an SspI site in RK. In the gL gene, a TTA-to-TAA substitution at codon position 59 resulted in the appearance of an additional ApoI cleavage site in RK. This substitution would change a leucine residue (JI) into a stop codon (RK). We made the hypothesis that the genetic changes at these critical positions supported the definition of alleles for the corresponding genes in the same sense as we had interpreted gB gene polymorphism previously.
FIG. 1.
Location and polymorphism sites of the p100, gB, MCP, and gL genes of the HHV-7 genome. The HHV-7 genome (144,861 bp long) is schematized at bottom, with the left and right terminal repeat sequences (TRL and TRR, respectively) and the internal repeat sequences (R1 and R2) noted. The different genetic sequences are presented at each codon mentioned for the four genes. The codon number of each gene corresponds to the published JI sequence (16). The five codon sequences of gB support the definition of six gB alleles as reported elsewhere (8).
Fifty HHV-7-positive samples from unrelated individuals previously analyzed with regard to gB alleles (8) were then studied at the critical positions of the p100, MCP, and gL genes by PCR and further genetic analysis of amplified products. Briefly, samples corresponding to 1 μg of DNA extracted from peripheral blood mononuclear cells were subjected to nested PCR as described previously (8) by using the primers indicated in Table 1. Amplified products were subjected to both nucleoside sequence determination and restriction endonuclease digestion as described previously (8). The digestion with the restriction endonucleases MboII, SspI, MunI, and ApoI was done in accordance with the manufacturers’ instructions (Boehringer, Mannheim, Germany; Ozyme, Beverly, Mass.); positive digestion controls were added in each experiment when the loss of a unique restriction site was suspected. In the case of the p100 gene, the two putative alleles corresponding to the presence of AAT and GAT at codon 721 were found in 21 (42%) and 29 (58%) of the 50 samples studied, respectively (Table 2). These two alleles were arbitrarily designated p100-A and p100-B. In the case of the MCP gene, four potential alleles, designated MCP-A, MCP-B, MCP-C, and MCP-D, were defined according to the four possible combinations of genetic changes at codons 763 and 774 (Table 2). Due to the limited amount of peripheral blood mononuclear cell DNA available for our study, only 36 samples were analyzed with regard to MCP alleles. The three alleles MCP-A, MCP-B, and MCP-C were found in 13 (36%), 3 (8%), and 20 (56%) of the samples studied, respectively. The putative allele MCP-D was not found, which suggested it was uncommon (<3%), if it existed. The same picture was observed for the putative gL allele B, which was not found in any of the 50 HHV-7-positive samples studied. The codon TAA at position 59 of the gL gene then appeared to be specific for RK and was no longer considered in our analysis. We concluded that the p100 and MCP genes exhibited polymorphism markers which permitted classification of the different HHV-7-positive samples into distinct groups as done previously with gB alleles.
TABLE 1.
Oligonucleotides used for PCR and sequencing
| Primer | Sequence (5′→3′) | Sense or antisense | Locationa | Use in nested PCR (type of gene) |
|---|---|---|---|---|
| MCP1 | AACTGTGTGGGGAACCATTGGTTGGA | Sense | 85442→85468 | Inner (MCP) |
| MCP2 | CTTGTGGTAGAGGAAAACCACGTC | Antisense | 84688→84713 | Inner (MCP) |
| MCP3 | GCAATAATCACAGCTGGTAACACTCCC | Sense | 85401→85429 | Outer (MCP) |
| MCP4 | TGCATGTGAGGGACCAACAGCTTC | Antisense | 85020→85043 | Outer (MCP) |
| p100.1 | GACATTACCCGAATAGGAAACG | Sense | 15901→15922 | Inner (p100) |
| p100.2 | GAAGTGTTCAATAATGCTCAACAG | Antisense | 18262→18285 | Inner (p100) |
| p100.3 | CTGTGTTTTAGACGACGTCTTC | Antisense | 15975→15996 | Outer (p100) |
| p100.4 | GCGACTTTCAATGACAACAACCTG | Sense | 16277→16301 | Outer (p100) |
| gL1 | CCAGCATCCACTGTAGTAATGCC | Antisense | 115926→115948 | Inner (gL) |
| gL2 | CCATATTTAAGTTCCTGGTG | Sense | 116801→116821 | Inner (gL) |
| gL3 | ACACACATTATCTCTACAGG | Antisense | 116361→116381 | Outer (gL) |
| gL4 | GTATACAACTTTATGGAACATG | Sense | 116776→116797 | Outer (gL) |
Reference strain JI.
TABLE 2.
Distribution of putative alleles of the p100, MCP, and gL genes among independent HHV-7-positive samples
| Type of gene and putative allele | Nucleotide sequence (codon position)a | Endonuclease cleavage site(s) | Reference strain | No. (%) of HHV-7-positive samples with indicated allele/no. of samples |
|---|---|---|---|---|
| p100 | ||||
| A | AAT (721) | JI | ||
| B | GAT (721) | MboII | RK | 29/50 (58) |
| MCP | ||||
| A | ATC (763) ACA (774) | MboII, MunI | JI | 13/36 (36)b |
| B | ATA (763) ACG (774) | SspI | RK | 3/36 (8) |
| C | ATA (763) ACA (774) | SspI, MunI | 20/36 (56) | |
| D | ATC (763) ACG (774) | MboII | 0/36 (0) | |
| gL | ||||
| A | TTA (59) | JI | 50/50 (100) | |
| B | TAA (59) | ApoI | RK | 0/50 (0) |
Based on the codon number of the published JI sequence.
Fourteen of the samples could not be studied with regard to the MCP alleles due to the lack of DNA.
Definition of two distinct HHV-7 prototype strains.
The recognition of the putative alleles of the p100, gB, and MCP genes, located at three distant positions of the HHV-7 genome, raised the question of whether the association between these alleles occurred at random. We then analyzed the frequency of allele combinations among the 36 samples for which the characterization of the three genes had been carried out (Table 3). Theoretically, when considering two, six, and three alleles for the p100, gB, and MCP genes respectively, 36 different combinations were possible. Only seven of these combinations (Co) were observed, and 75% of the samples corresponded to two of them, designated provisionally Co1 and Co2. The reference strain JI was defined as Co2, as were our reference isolate, IM (17), and the original HHV-7-positive sample which was the source of IM. The reference strain RK corresponded to an eighth combination (Co8) which was not observed in the group of human samples we investigated. In accordance with the nonrandom distribution of allele combinations, statistical analyses showed a significant association between the alleles gB-C, p100-A, and MCP-C on one hand, and the alleles gB-F, p100-B, and MCP-A on the other (P < 0.001; chi-square test). These results supported the idea that Co1 and Co2 were the genetic signatures of two predominant prototype variants of HHV-7 and that less frequent allele combinations might correspond to HHV-7 strains which have been derived from prototype variants by point mutation and/or recombination events.
TABLE 3.
Distribution of allele combinations in HHV-7-positive samples
| Sample or allele combination | Allele at gene
|
No. (%) of samples with indicated combination | ||
|---|---|---|---|---|
| p100 | gBa | MCP | ||
| Reference strain | ||||
| JI | A | F | A | NAb |
| RK | B | C | B | NA |
| IM | A | F | A | NA |
| Allele combination | ||||
| Co1 | B | C | C | 16 (44) |
| Co2 | A | F | A | 11 (31) |
| Co3 | B | F | C | 3 (8) |
| Co4 | B | C | A | 2 (6) |
| Co5 | B | D | B | 2 (6) |
| Co6 | A | C | C | 1 (3) |
| Co7 | A | F | C | 1 (3) |
| Co8 | B | C | B | 0 |
The results concerning the gB alleles were partly published elsewhere (8).
NA, not applicable.
The genome of HHV-7 appears to be highly conserved, as reflected by the high degree of nucleotide sequence homology between the unrelated strains JI and RK (15, 16). This conservation has been previously shown for the gB gene with 99.8% nucleotide sequence homology (8) and was confirmed here for three other genes (p100, MCP, and gL) distantly located on the viral genome. However, albeit limited, the polymorphism of the gB gene permitted the definition of alleles, this term designating stable associations of genetic markers within this gene. We have now extended this concept to the p100 and MCP genes: one critical codon in the former gene and two critical codons in the latter permitted the classification of HHV-7-positive samples into two distinct groups in the case of the p100 gene and three distinct groups in the case of the MCP gene. The two groups based on the polymorphism of codon 721 of p100 differed from each other by the amino acid residue (asparagine or aspartic acid) corresponding to this codon. In contrast, the three groups based on the polymorphism of codons 763 and 774 of MCP did not differ from each other with regard to the amino acid sequence at the corresponding position. This was also the case for the gB gene alleles (8), and we have shown here that gB gene polymorphism, silent at the protein level, was apparently not associated with differences in either transcription or translation processes. Given that the effects of genetic polymorphism on virus phenotypes are very modest, if they exist, the different alleles we have characterized may be considered simply as stable genetic entities transmitted without apparent selection pressure. What we presently know about HHV-7 epidemiology and pathogenicity fits this scenario. HHV-7 is transmitted early in life via saliva, inducing an asymptomatic or poorly symptomatic primary infection which results in a lifelong chronic infection with apparently no major associated disease. This ubiquitous virus is therefore assumed to be serially propagated in human communities with a very high conservation of its genetic content through generations. This genetic stability implies that not only individual alleles but also allele combinations would be maintained through serial transmission. In agreement with this view, we have described in the present paper two preferential associations of p100, gB, and MCP alleles, representing 44 and 31% of the HHV-7-positive samples studied, which, in our opinion, may support the concept of prototype HHV-7 variants. These two allele combinations may represent two major prototype strains that have been propagated independently of each other. Of note, Co1 contains the allele gB-C, which has been found more frequently in African and Caribbean subjects, while Co2 contains the allele gB-F, which is more frequent in European subjects (8). It is tempting to anticipate that the two putative variants are distributed as gB alleles, according to the geographical origin of the samples. This conclusion, which requires a definite demonstration by means of an ongoing larger study, points out the interest in HHV-7 variants as markers for studying population movements and genetics, as previously concluded from the study of gB alleles. Two other problems remain unsolved: (i) the possibility of different biological behaviors of the two HHV-7 variants due to subtle genetic differences and (ii) the genetic mechanisms by which minor variants (at least five allele combinations representing 25% of the samples studied) have been derived from the two major prototype variants. Recombination after a mixed infection appears to be a likely hypothesis, keeping in mind that the infection with two distinct strains of HHV-7 may be observed in vivo (8). However, the occurrence of rare point mutations at critical codons still fits the very high degree of conservation of the HHV-7 genome and cannot be ruled out as a factor in genetic evolution.
Acknowledgments
This work was supported in part by the Association Claude Bernard, the Action Concertée Coordonnée des Sciences du Vivant of the French Ministry of Research, and MRTC research grant no. 97-5-12172 to M.F.
REFERENCES
- 1.Berneman Z N, Gallo R C, Ablashi D V, Frenkel N, Katsafanas G, Kramarsky B, Brus I. Human herpesvirus 7 (HHV-7) strain JI: independent confirmation of HHV-7. J Infect Dis. 1992;166:690–691. doi: 10.1093/infdis/166.3.690. [DOI] [PubMed] [Google Scholar]
- 2.Berneman Z N, Torelli G, Luppi M, Jarrett R F. Absence of a directly causative role for human herpesvirus 7 in human lymphoma and a review of human herpesvirus 6 in human malignancy. Ann Hematol. 1998;77:275–278. doi: 10.1007/s002770050457. [DOI] [PubMed] [Google Scholar]
- 3.Berneman Z N, Ablashi D V, Lee G, Eger-Fletcher M, Reitz M S, Hung C L, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black J B, Inoue N, Kite-Powell K, Zaki S, Pellett P E. Frequent isolation of human herpesvirus 7 from saliva. Virus Res. 1993;29:91–98. doi: 10.1016/0168-1702(93)90128-a. [DOI] [PubMed] [Google Scholar]
- 5.Clark D A, Freeland J M L, Markie P L K, Jarrett R F, Onions D E. Prevalence of antibody to human herpesvirus 7 by age. J Infect Dis. 1993;168:251–252. doi: 10.1093/infdis/168.1.251. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez G, Black J B, Stamey F R, Inoue N, Pellet P E. Physical and genetic map of the human herpesvirus 7 strain SB genome. Arch Virol. 1996;141:2387–2408. doi: 10.1007/BF01718639. [DOI] [PubMed] [Google Scholar]
- 7.Drago F, Ranieri E, Rebora A. Pityriasis rosea and herpesvirus 7: action or interaction? Dermatology. 1998;197:275. [PubMed] [Google Scholar]
- 8.Franti M, Aubin J-T, Poirel L, Gautheret-Dejean A, Candotti D, Huraux J-M, Agut H. Definition and distribution analysis of glycoprotein B gene alleles of human herpesvirus 7. J Virol. 1998;72:8725–8730. doi: 10.1128/jvi.72.11.8725-8730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frenkel N, Schirmer E C, Wyatt L S, Katsafanas G, Roffman E, Danovich R M, June C H. Isolation of a new herpesvirus from human CD4+ T cells. Proc Natl Acad Sci USA. 1990;87:748–752. doi: 10.1073/pnas.87.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautheret A, Aubin J-T, Poirel L, Huraux J-M, Nicolas J-C, Rozenbaum W, Agut H. Detection of human Betaherpesvirinae in saliva and urine from immunocompromised and immunocompetent subjects. J Clin Microbiol. 1997;35:1600–1603. doi: 10.1128/jcm.35.6.1600-1603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata A, Mukai T, Isegawa Y, Yamanishi K. Identification and analysis of glycoprotein B of human herpesvirus 7. Virus Res. 1996;46:125–137. doi: 10.1016/s0168-1702(96)01395-0. [DOI] [PubMed] [Google Scholar]
- 12.Kasolo F C, Mpabalwani E, Gompels U A. Infection with AIDS-related herpesviruses in human immunodeficiency virus-negative infants and endemic childhood Kaposi’s sarcoma in Africa. J Gen Virol. 1997;78:847–855. doi: 10.1099/0022-1317-78-4-847. [DOI] [PubMed] [Google Scholar]
- 13.Lusso P, Secchiero P, Crowley R W, Garzino-Demo A, Berneman Z N, Gallo R C. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci USA. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malet I, Wychowski C, Huraux J M, Agut H, Cahour A. Yellow fever 5′ noncoding region as a potential element to improve hepatitis C virus production through modification of translational control. Biochem Biophys Res Co. 1998;253:257–264. doi: 10.1006/bbrc.1998.9740. [DOI] [PubMed] [Google Scholar]
- 15.Megaw A G, Rapaport D, Avidor B, Frenkel N, Davison A J. The DNA sequence of the RK strain of human herpesvirus 7. Virology. 1998;244:119–132. doi: 10.1006/viro.1998.9105. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel P, Aubin J T, Gautheret A, Malet I, Huraux J M, Agut H. Use of inverse polymerase chain reaction to characterize a novel human herpesvirus 7 isolate. J Virol Methods. 1997;64:197–203. doi: 10.1016/s0166-0934(96)02167-2. [DOI] [PubMed] [Google Scholar]
- 18.Portolani M, Cermelli C, Mirandola P, DiLuca D. Isolation of human herpesvirus 7 from an infant with febrile syndrome. J Med Virol. 1995;45:282–283. doi: 10.1002/jmv.1890450307. [DOI] [PubMed] [Google Scholar]
- 19.Sato A, Nakagawa M, Nishizawa K, Narita T, Nishikawa R, Yamada A, Ishizaki T. Thrombocytopenia after human herpesvirus-7 infection in a patient with DiGeorge syndrome. Pediatr Hematol Oncol. 1999;21:171–172. doi: 10.1097/00043426-199903000-00018. [DOI] [PubMed] [Google Scholar]
- 20.Secchiero P, Sun D, de Vico A L, Crowley R W, Reitz M S, Jr, Zauli G, Lusso P, Gallo R C. Role of the extracellular domain of human herpesvirus 7 glycoprotein B in virus binding to cell surface heparan sulfate proteoglycans. J Virol. 1997;71:4571–4580. doi: 10.1128/jvi.71.6.4571-4580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Secchiero P, Bonino L D, Lusso P, Abele M C, Reato G, Kerim S, Palestro G, Zauli G, Valente G. Human herpesvirus type 7 in Hodgkin’s disease. Br J Haematol. 1998;101:492–499. doi: 10.1046/j.1365-2141.1998.00718.x. [DOI] [PubMed] [Google Scholar]
- 22.Sugaya S, Fujita K, Kikuchi A, Ueda H, Takakuwa K, Kodama S, Tanaka K. Inhibition of tumor growth by direct intratumoral gene transfer of herpes simplex virus thymidine kinase gene with DNA-liposome complexes. Hum Gene Ther. 1996;7:223–230. doi: 10.1089/hum.1996.7.2-223. [DOI] [PubMed] [Google Scholar]
- 23.Van den Berg J S, van Zeijl J H, Rotteveel J J, Melchers W J, Gabreels F J, Galama J M. Neuroinvasion by human herpesvirus type 7 in a case of exanthem subitum with severe neurologic manifestations. Neurology. 1999;52:1077–1079. doi: 10.1212/wnl.52.5.1077. [DOI] [PubMed] [Google Scholar]
- 24.Wallace H L, II, Natelson B, Gause W, Hay J. Human herpesviruses in chronic fatigue syndrome. Clin Diagn Lab Immunol. 1999;6:216–223. doi: 10.1128/cdli.6.2.216-223.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilborn F, Schmidt C A, Lorenz F, Peng R, Gelderblom H, Huhn D, Siegert W. Human herpesvirus type 7 in blood donors: detection by the polymerase chain reaction. J Med Virol. 1995;47:65–69. doi: 10.1002/jmv.1890470113. [DOI] [PubMed] [Google Scholar]
- 26.Wyatt L S, Frenkel N. Human herpesvirus 7 is a constitutive inhabitant of adult human saliva. J Virol. 1992;66:3206–3209. doi: 10.1128/jvi.66.5.3206-3209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyatt L S, Rodriguez W J, Balachandran N, Frenkel N. Human herpesvirus 7: antigenic properties and prevalence in children and adults. J Virol. 1991;65:6260–6265. doi: 10.1128/jvi.65.11.6260-6265.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]