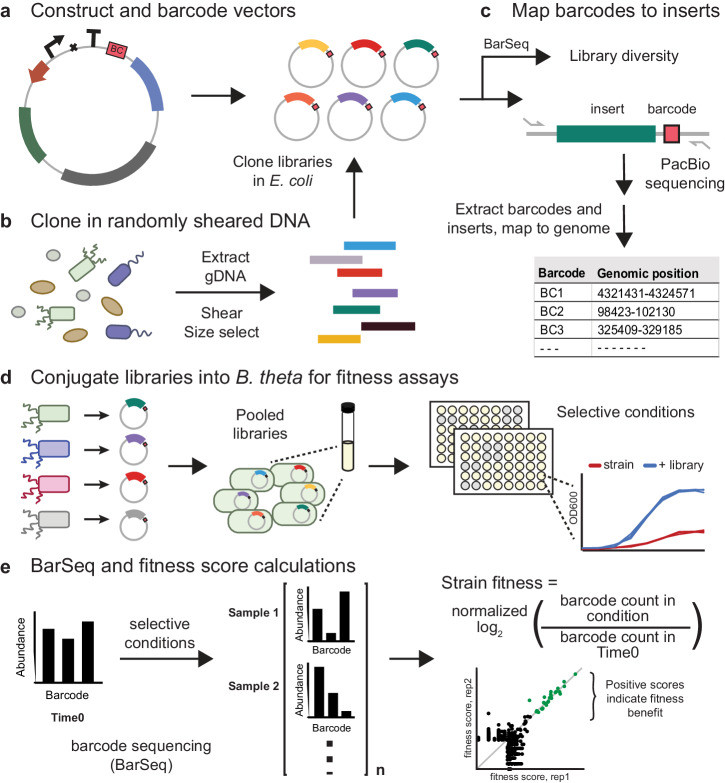
Fig. 1. The Boba-seq workflow for high-throughput functional screens.
a Clone DNA barcodes into vectors that replicate in the final overexpression host. b Extract DNA, shear, size-select, and insert into barcoded vectors using an E. coli cloning strain. c Quantify library diversity using BarSeq. Map barcodes to inserts using long-read sequencing technology and generate a barcode-to-insert mapping table. d Conjugate mapped libraries into the overexpression strain for fitness screens. Libraries can be pooled to increase the genetic diversity assayed. A range of selective conditions can be used to determine fitness differences across strains, such as carbon utilization and stress tolerance. e Quantify library composition before and after growth on selective conditions using BarSeq. A fitness score is calculated for each barcode or strain. Significantly high fitness scores in both replicates point to putative beneficial genes in the condition.