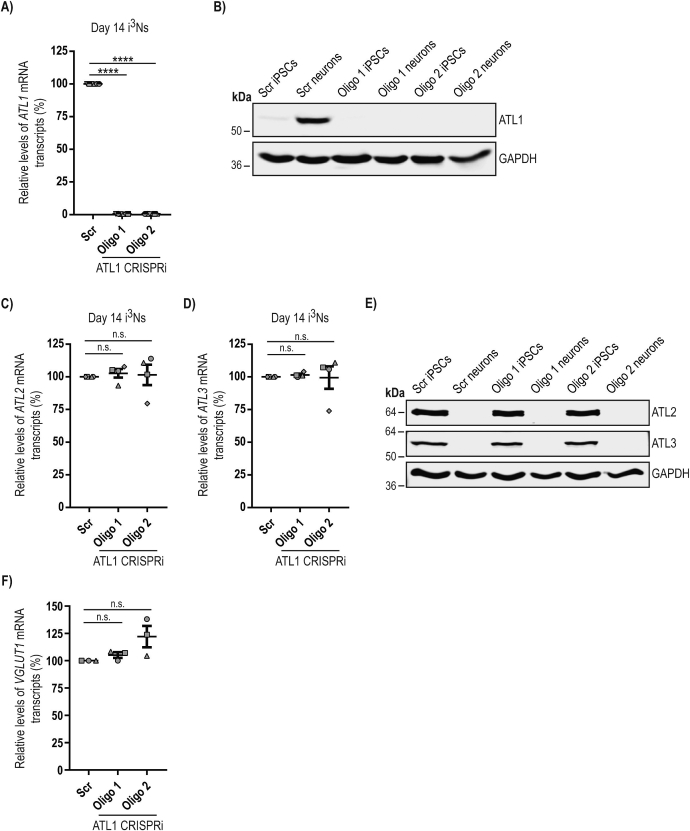
Fig. 2.
Generation and characterisation of human neurons lacking atlastin-1.
A) Relative mRNA abundance of ATL1 transcript in day 14 CRISPRi-i3Ns (right) expressing a scrambled sgRNA, or ATL1-targeting sgRNAs oligo 1 or oligo 2, measured by qPCR. Data are displayed relative to the scrambled value. Error bars show mean values +/− SEM of 8 i3Ns biological repeats, each carried out in triplicate. B) Immunoblots showing the abundance of atlastin-1 protein in iPSCs or day 14 day i3Ns expressing the sgRNAs indicated. GAPDH blotting serves as a loading control. C and D) Relative mRNA abundance of ATL2 (C) or ATL3 (D) transcript in day 14 i3Ns expressing the sgRNAs shown, measured by qPCR. Data are displayed relative to the scrambled value. Error bars show mean values +/− SEM of 4 biological repeats, each carried out in triplicate. In all qPCR experiments ATL transcript levels were normalised against the ACTB reference gene. E) Representative immunoblots showing the abundance of atlastin proteins in iPSCs or day 14 i3Ns generated from the CRISPRi lines shown. GAPDH blotting serves as a loading control. F) Relative abundance of VGLUT1 in day 14 i3Ns expressing the sgRNAs shown, measured by qPCR. Data are displayed relative to the scrambled value. Error bars show mean values +/− SEM of 3 biological repeats. VGLUT1 transcript levels were normalised against the GAPDH reference gene. In A), C), D) and F) statistical comparisons were carried out with one-way ANOVA with Dunnett's test for multiple comparisons, n.s., p > 0.05; ****, p < 0.0001.