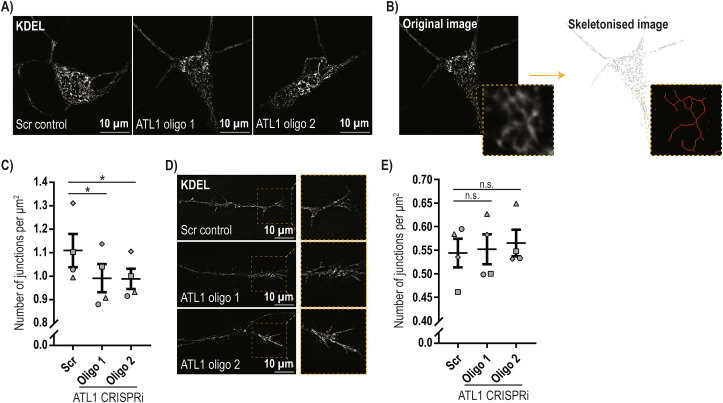
Fig. 4.
Depletion of atlastin-1 causes reduced ER three-way junction abundance in human neurons.
A) i3Ns from the CRISPRi lines indicated were transduced with a doxycycline-inducible mEmerald-KDEL lentiviral construct and treated with doxycycline for three days from day 11 of differentiation. They were then imaged live with a Zeiss Elyra 7 microscope with Lattice SIM2 super-resolution on day 14. Representative images are shown. B) ER morphology was analysed using Image J software. Images were analysed blind. Each image was thresholded to select the KDEL signal and then skeletonised to show the network simplified into single pixel wide lines. The zoomed-in skeleton area on the bottom right shows representative skeleton labelling post-analysis. End-point voxels (blue) are pixels with only 1 neighbouring labelled pixel; junction voxels in purple are pixels with >2 neighbours; and slab voxels in orange have exactly 2 neighbours. A neighbouring group of junction voxels is counted as one junction. C) Junctions were counted in neuronal bodies and the number of junctions was normalised to the cell area. Four neuronal differentiation biological repeats were analysed with at least 25 neurons per genotype in each repeat, with the exception of one repeat for oligo 1 which was based on 4 neurons. Bars show mean +/− SEM. D) A similar analysis of ER morphology to that shown in B) was carried out in growth cone regions. The images show representative micrographs of growth cones labelled with mEmerald-KDEL in the i3N lines indicated. E) Four neuronal differentiation biological repeats were analysed with a minimum of 6 (mean of 27.7) growth cones per genotype analysed in each repeat. The mean number of junctions normalised to the growth cone area was plotted +/− SEM. In C) and E) statistical comparisons were performed with repeated measure one-way ANOVA, with Dunnett's correction for multiple testing. n.s., p > 0.05; *, p < 0.05.