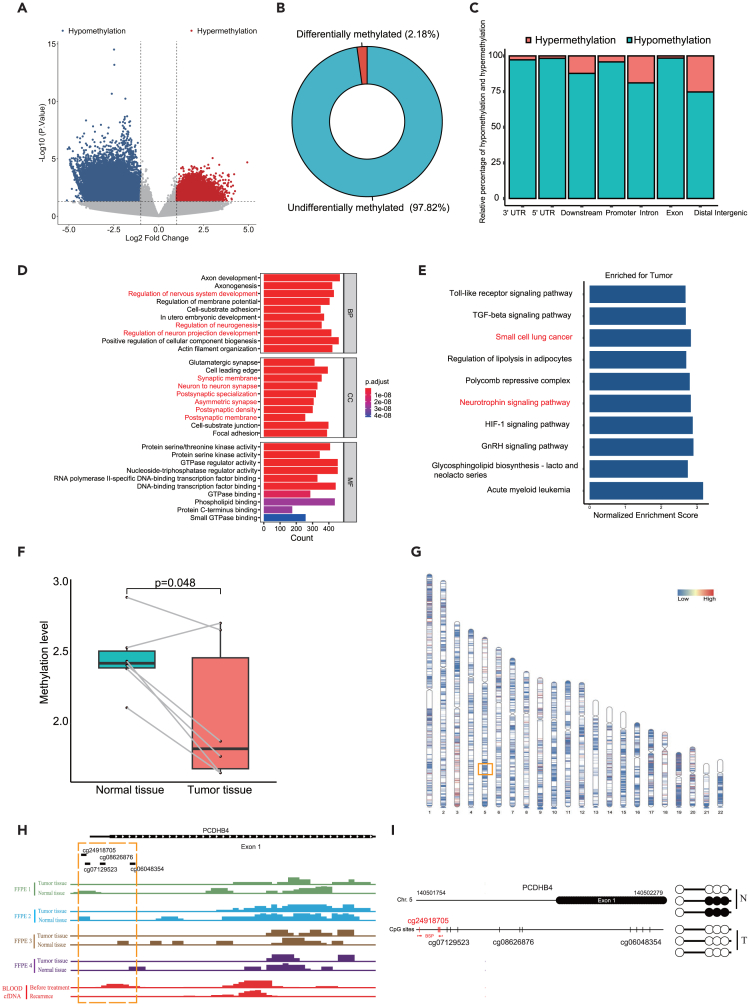
Figure 5.
The validation of hypomethylation of the PCDHB4 gene in SCLC specimens by MeDIP-seq and BSP
(A) Volcano plot of differentially methylated sites between SCLC tumors and adjacent normal tissues by MeDIP-seq.
(B) The proportion of differentially methylated sites in all detected methylated sites.
(C) The category of genomic locations for hypermethylated or hypomethylated sites.
(D) GO analysis for the differentially methylation genes. Top 10 terms are shown. Neuron-associated pathways are marked in red.
(E) Bar plot of GSEA based on the differentially methylation genes.
(F) Boxplot showing the methylation level within the PCDHB4 promoter in SCLC tumors and adjacent normal tissues.
(G) Distribution of differentially methylated sites on chromosomes. The red area represents the hypermethylated region, and the blue area represents the hypomethylated region. The PCDHB4 gene locates in the orange box.
(H) The peak maps of methylation with PCDHB4 promoter by MeDIP-seq. DNA methylation was detected in SCLC tumors and adjacent normal tissues (FFPE 1, FFPE 2, FFPE 3, and FFPE 4) by MeDIP-seq. cfDNA methylation of an SCLC patient (before treatment and after recurrence) was detected by MeDIP-seq.
(I) BSP analysis of the CpG site cg24918705 in SCLC tumor and adjacent normal tissue.