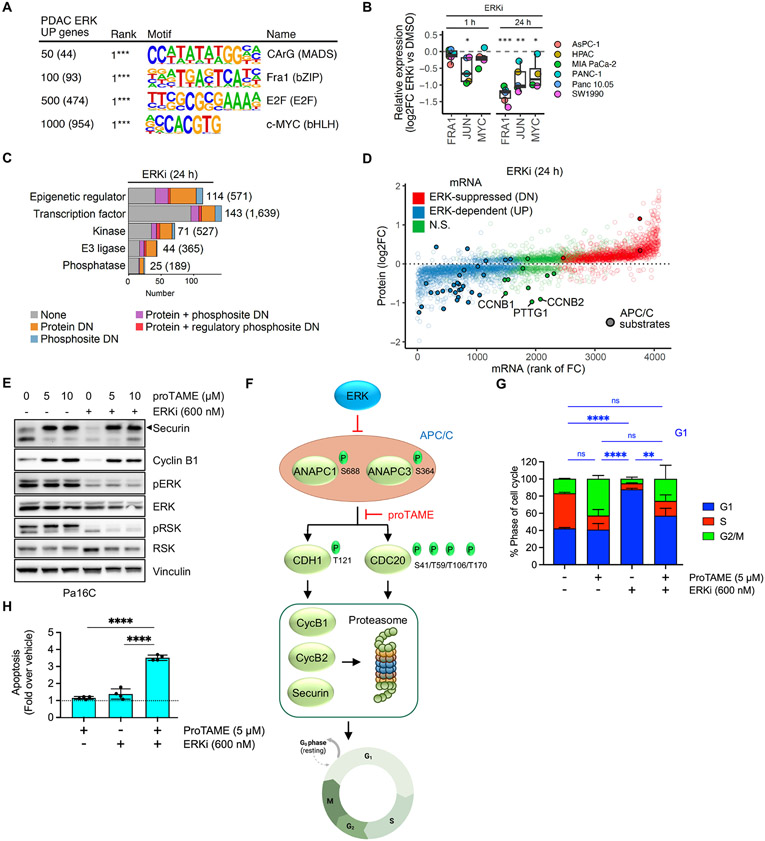
Fig. 3. Integration of transcriptomic, proteomic, and phosphosite activity data reveals multiple ERK dependent cell cycle control mechanisms.
(A) Supervised gene promoter motif enrichment analysis for different cutoffs of the top PDAC ERK UP genes. Numbers in parentheses indicate number of genes from each subset used in the analysis. (B) Protein abundance of AP-1 transcription factor components FRA1 and JUN, and of MYC, after 1 and 24 hours of ERKi, (SCH772984, 1000 nM) treatment across six PDAC cell lines, was determined by LC-MS2. (C) Categorization of proteins dependent on ERK for expression and/or phosphosite activity into five main functional groups. Numbers in parentheses indicate total number encoded in human genome. (D) Comparison of change in protein abundance (y-axis) with change in mRNA expression (x-axis) following ERKi treatment (24 hours) for proteins with significant changes (log2FC > 0.2 & adj. p-val. < 0.05). APC/C substrates are indicated by filled circles. (E) Immunoblot analysis of APC/C target proteins securin and cyclin B1 as well as ERK activity readouts pERK and pRSK following treatment with proTAME (APC/Ci) and/or ERKi in Pa16C cells. Images are representative of 2-3 biological replicates. (F) Schematic for ERK inhibition of APC/C E3 ligase function and degradation of mitotic regulators securin and cyclin B1/2. (G) Cell cycle distribution analysis of Pa16C cells treated with APC/Ci and/or ERKi for 24 hours (n = 3, each condition). (H) Apoptosis induction relative to vehicle control in Pa16C cells treated with APC/Ci and/or ERKi for 3 days (n = 4, each condition). (G-H), error bars represent +/− 1 S.D. All statistical comparisons, ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.