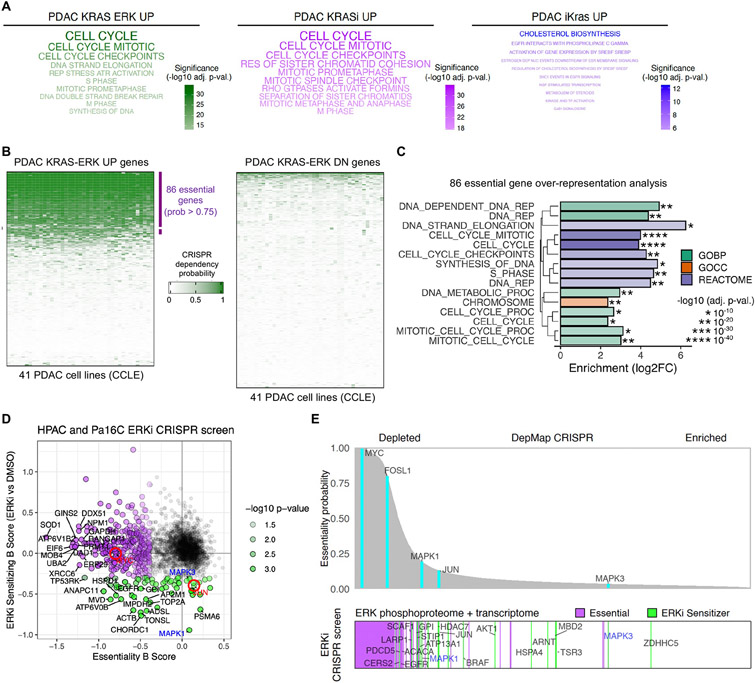
Fig. 4. KRAS-ERK dependent genes are essential for cell proliferation in PDAC.
(A) Over-representation analysis for REACTOME terms in three KRAS signatures: PDAC KRAS-ERK UP, PDAC KRASi UP, and PDAC iKras UP. Top 10 terms are shown. (B) Comparison of CRISPR dependency probabilities to gauge essentiality of genes in PDAC KRAS-ERK UP/DN signatures across 41 KRAS-mutant PDAC cell lines from CCLE. (C) Over-representation analysis for PDAC KRAS-ERK UP essential genes using KEGG, GO, and REACTOME. BP, biological process; CC, cellular component. (D) CRISPR drop-out screen using sgRNA library generated from ERK-dependent phosphoproteins and transcripts. Beta scores calculated with MAGeCK. Red circles highlight MYC and JUN. (E) Comparison of genes highlighted in (D) (bottom) with essentiality probabilities averaged across the 41 KRAS-mutant PDAC cell lines in DepMap (top).