Abstract
Copy number variations (CNVs) which include deletions, duplications, inversions, translocations, and other forms of chromosomal re-arrangements are common to human cancers. In this report we investigated the pattern of these variations with the goal of understanding whether there exist specific cancer signatures. We used re-arrangement endpoint data deposited on the Catalogue of Somatic Mutations in Cancers (COSMIC) for our analysis. Indeed, we find that human cancers are characterized by specific patterns of chromosome rearrangements endpoints which in turn result in cancer specific CNVs. A review of the literature reveals tissue specific mutations which either drive these CNVs or appear as a consequence of CNVs because they confer an advantage to the cancer cell. We also identify several rearrangement endpoints hotspots that were not previously reported. Our analysis suggests that in addition to local chromosomal architecture, CNVs are driven by the internal cellular or nuclear physiology of each cancer tissue.
Keywords: Copy number variations, DNA double strand breaks, Cancer, Chromosomal rearrangements
1. Introduction
Cancers are characterized by accumulation of mutations in coding and regulatory gene regions, changes in gene expression, and structural genomic rearrangements [1]. Genomic rearrangements which include large deletions, duplications, insertions, chromosomal translocations as well as chromosome gain or loss caused by mitotic non-disjunction lead to copy number variations (CNVs) [2]. CNVs are thought to promote cellular transformation and immortalization by disrupting tumor suppressor genes (e.g. loss of heterozygosity caused by chromosome loss or deletion), increasing the copy number of oncogenes (insertion, duplication, chromosome gain) or altering gene expression (translocations) [3].
Cancer genetics is replete with models of how CNVs contribute to cellular immortalization [1]. For example, in many cancers, several key tumor suppressors (CDKN2A, TP53, RB1, PTEN, etc.) are inactivated by homozygous deletion [4–9] while certain proto-oncogenes are modified by amplification (MET, BRAF, etc.) [10–13]. Translocations can also produce changes in gene expression. The most famous is the Philadelphia chromosome which results from a reciprocal translocation between chromosomes 9 and 22, t(9;22)(q34;q11), most often in chronic myeloid leukemias but also in some other forms of blood cancers [14–16]. This translocation creates the BCR-ABL1 fusion which results in constitutively active ABL1 tyrosine kinase [17]. More examples include chromosome 17 loss/deletion [18] as well as others reviewed elsewhere [9].
Recent studies predict that many chromosomal re-arrangements occur during a brief period of time (one or two cell cycles) by processes known as chromothripsis [19–21] or break fusion bridge (BFB) cycles [22]. In this process, chromosomes are virtually shattered and rejoined but not with the same genomic organization as in quiescent cells, thus producing a massive genomic rearrangement event.
Unlike whole chromosomal copy number changes which are generally due to mitotic non-disjunction or endo replication, the major driver of chromosomal rearrangements is incorrectly repaired DNA double strand breaks (DSBs) [23,24]. DSBs can be generated by exogenous factors such as chemicals or radiation as well as endogenous processes such as DNA replication and transcription [25]. Regardless of how they happen, both types of breaks are repaired by the same genetic mechanisms which in human cells can be generally subdivided into homologous recombination (HR) and non-homologous end joining (NHEJ) [26]. Sub-pathways have been identified in both mechanisms. HR functions mainly during S-phase and G2 because it requires a homologous template which is usually found on the sister chromatid. Conversely, NHEJ is active during G1 but also contributes to repair during the other cell stages [27].
It is virtually impossible to determine precisely how many DSBs occur in human cancer genomes because we can only analyze those breaks that have been repaired incorrectly. Nevertheless, by comparing analyzed cancer genomes with the reference genome, it is possible to identify segments that have been reorganized, thus inferring where errors in DNA damage repair have occurred. Several studies have previously identified and mapped CNVs in human genomes [3,28–32]. A recent report has also characterized structural variations using data from Pan-Cancer Analysis of Whole Genomes and they identified certain cancer specific signatures [33]. In this report we provide a review of the literature and analyze all genomic rearrangement endpoints from independent studies deposited on the Catalogue of Somatic Mutations in Cancers (COSMIC) and generated tissue specific maps.
2. Materials and methods
2.1. Data acquisition
An excel file with the structural genomic rearrangements breakpoints was downloaded from COSMIC [34]. This file contains data on 16 cancers and some that are not specified (Supplementary Fig. S1). For each cancer, data was acquired from multiple samples except for the upper aerodigestive tract that only has 2 samples. Several aberrations are reported within each sample type (Supplementary Table 1). References are provided for samples belonging to published studies (Supplementary Table 2).
2.2. Graphs and figures
Graphs were created in either SPSS or Excel. The COSMIC data lists chromosomal coordinates for “Location From” and “Location To” indicating the first breakpoint “from” and second breakpoint “to” (Fig. 1A). For each location, the minimum and maximum detected coordinates are listed (e.g. location from minimum, location from maximum, location to minimum, and location to maximum). The minima and maxima represent the resolution level within a range of base pairs. For some samples, the minima and maxima coordinates are identical because they are generated by whole genome sequencing while for others which were identified using microarray analysis this position is given at the resolution of the probe. The graph in Fig. 1C represents “location from” minimum (first breakpoint) and “location to” minimum (second breakpoint). The bottom graphs represent all breakpoints (first and second). For Fig. 1D and E the size of the fragments was generated by subtracting “location from minimum” from “location to maximum”. Because inversions produce a negative number, all values were multiplied by -1. For the data in Fig. 2, coordinates for location from minimum and location to maximum were combined and graphed together. The goal was to generate a graph with all the breaks (both 1st and 2nd break).
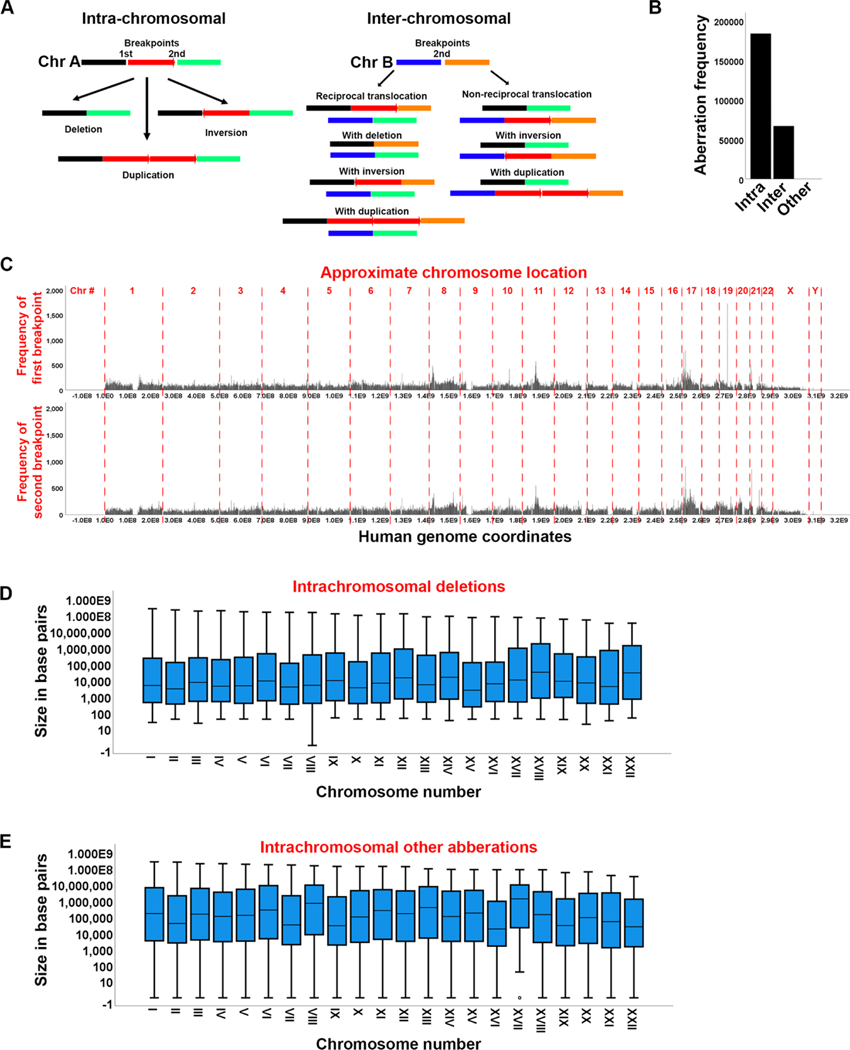
Fig. 1. Distribution of chromosomal aberration endpoints in human cancers.
A. Diagrams showing some of the possible intra- and inter-chromosomal aberrations. An aberration is defined as improper connection of two breakpoints (1st and 2nd). This can result in intra-chromosomal deletion, inversion or duplication and various inter-chromosomal duplications. B. Frequency of intra- and inter-chromosomal aberrations. “Other” represents complex or not specified forms of aberrations. C. Genome density of the two aberration endpoints (breakpoints) leading to the aberrations in A. The approximate chromosome location is indicated on top. D, E. Size of intrachromosomal deletions (D) or other intra-chromosomal aberrations (E) in base pairs.
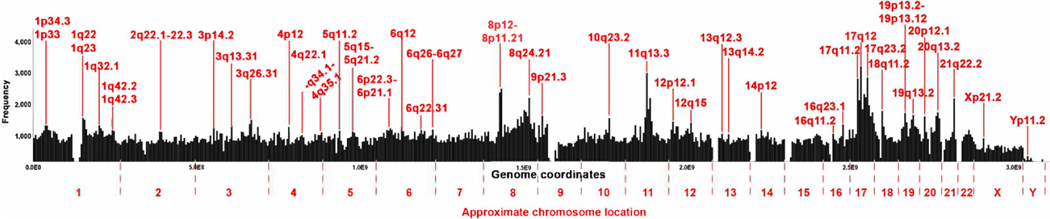
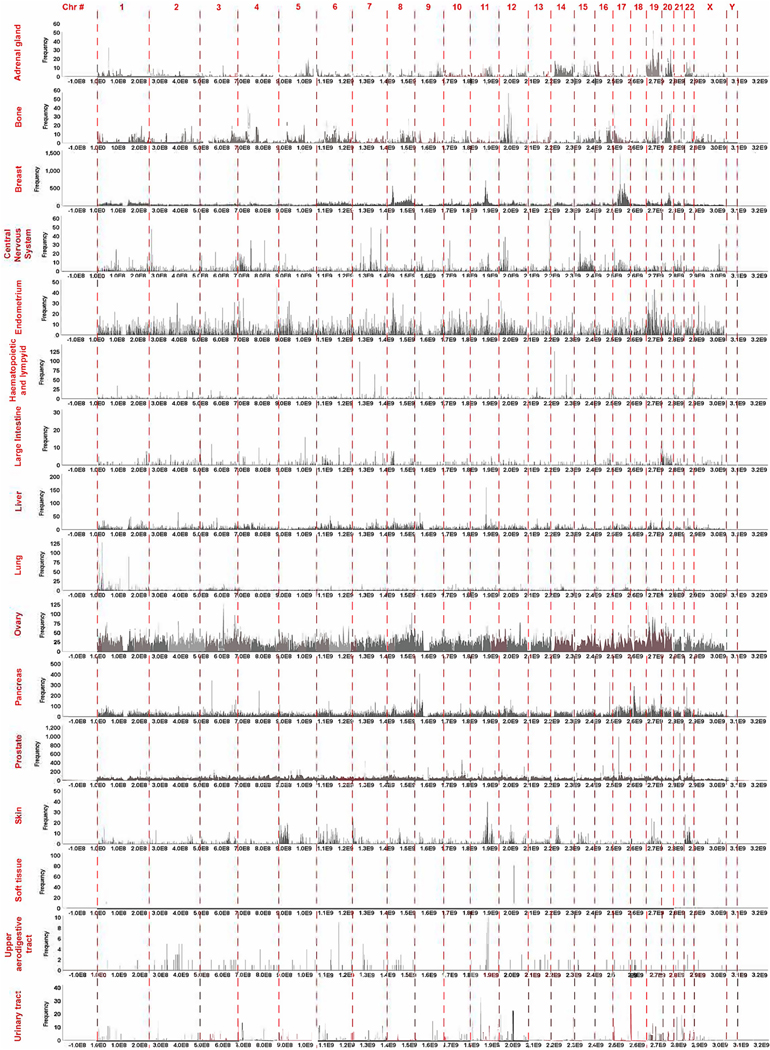
Fig. 2. Aberration endpoint (breakpoints) density in the human cancer genomes.
The frequency of the combined first and second aberration endpoints is shown. For each chromosome the location of the segments characterized by an increase in break density is shown. Each bar represents one million base pairs. The genome coordinates are given in base pairs and the approximate chromosome location is also indicated.
2.3. Computational analysis
Supplementary Fig. 4 shows the matrices representing the frequency of the aberrations in each cancer type. The matrices are derived based on the computational analysis implemented in Python 3 and described as follows:
The chromosomal aberrations from Cosmic are imported for each cancer type, and relevant columns are extracted for the two breakpoints (“Location From” and “Location To” ). A frequency matrix, , which is a square matrix of size 24 (for 24 chromosomes) is computed. This matrix represents the frequency of the chromosomal aberrations, and each element of the matrix represents the total number of chromosomal aberrations between chromosomes and . Due to the different lengths of the chromosomes in human genome, a new matrix is defined by normalization of . Since an element of the matrix is derived from two chromosomes and , the corresponding normalized element, of matrix is derived as follows:
| (1) |
| (2) |
| (3) |
where and are the length of chromosomes and , respectively.
To scale elements of in the range of [0,1], all elements are divided by the maximum element in as follows:
| (4) |
is the normalized frequency matrix representing the frequency of the intrachromosomal aberrations (diagonal elements), and inter-chromosomal aberrations (none-diagonal elements). Fig. 4A is created using normalized frequency matrices.
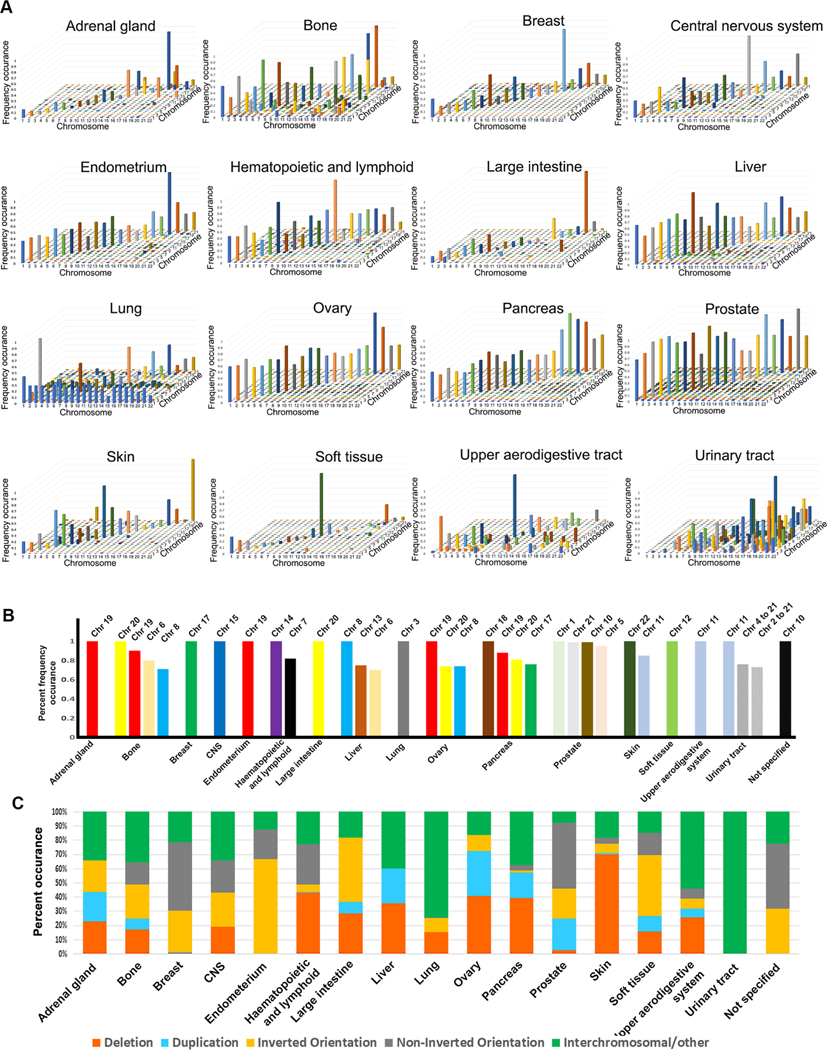
Fig. 4. Aberration endpoint signatures in human cancers.
A. Graphs of concordance matrices to map aberration endpoints density. The X-axis represents one aberration endpoint (1st breakpoint) while the Z-axis represents the other (2nd breakpoint). The Y-axis represents incidence frequency. B. Summary of concordance matrices to map chromosome specific aberration endpoints (breaks) in different cancers (see Supplementary Fig. S5). This graph shows only those chromosomes with a frequency occurrence of 0.7 or above. Complete data is shown in Supplementary Fig. S5. C. Distribution of the several types of chromosomal aberrations by cancer type. “Non-inverted orientation” represents unknown or uncharacterized.
The graphs in Fig. 5 are created using the Circos tool [35] for visual representation of chromosomal aberrations. The files are appropriately formatted for input to the Circos.
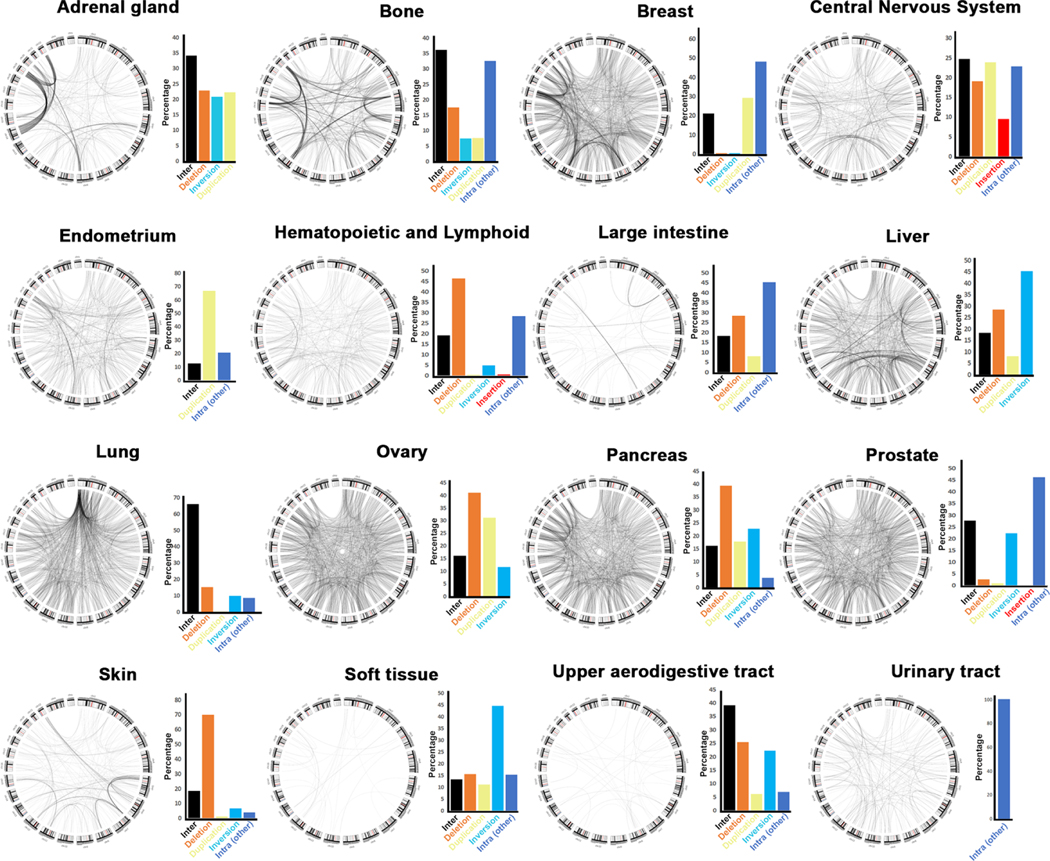
Fig. 5. Signature inter-chromosomal aberrations in human cancers.
Circos plots showing inter-chromosomal translocations by cancer type. Next to each graph, the percent of intra-chromosomal aberrations is shown for comparison. These circus plots represent all re-arrangements reported on COSMIC (e.g., from all publications). Publications where these data were first reported may have graphed subsets of these rearrangements. Some data were not previously published. Please see Table 1 for complete references for all these data.
The spider graphs in Supplementary Fig. 6 for intra-chromosomal aberrations are created in Python 3 according to the following computational analysis:
The different types of aberrations (deletion, insertion, inversion, duplication, with inverted orientation, non-inverted orientation, and unknown type) are filtered for each cancer type. The graphs are created for each aberration type by developing two functions: 1) function “equation_constant” that solves a system of linear equations using a NumPy function (numpy.linalg.solve), given three coordinates for the first, mid, and end points for each aberration. 2) A function “create_curve” that creates two arrays of , for generating , coordinates of the points between two breakpoints, as follows:
| (5) |
| (6) |
where , , and are the coefficients of the system of linear equations resulted from “equation constant” function, is the coordinate of the first breakpoint, and is defined as (we used as the total number of points). Finally, the spider plots were created based on and vectors.
3. Results
Chromosomal aberrations can be generally subdivided into intra-chromosomal and inter-chromosomal CNVs (Fig. 1A). Most chromosomal re-arrangements are hypothesized to require two breaks though one break is sometimes sufficient [36]. For example, a chromosomal deletion can occur if two breaks on either side of the red segment (Fig. 1A) are improperly repaired to exclude the red segment while an inversion or duplication can also be easily envisioned as resulting from improper repair of two breaks. A deletion may also occur from bidirectional resection of both ends of a break followed by ligation as in the case of single strand annealing [37]. This usually happens if the break is found between two direct repeats. Inter-chromosomal re-arrangements can occur if a break on one chromosome is improperly joined with a break on another chromosome. Most genomic re-arrangements are intra-chromosomal (deletions and duplications, inversions) which agrees with previous findings [33] (Fig. 1B). Deletions, duplications, and inversion in non-coding regions that do not contain genes that drive cellular transformation are unlikely to significantly affect cancer progression. However, the role of these deletions in promoting cellular transformation and immortalization is debated [9] and discussed later. Previous findings have also reported that deletions are frequent in late replicating origins while duplications correlated with early replicating origins [33]. About 25–30 % of aberrations are inter-chromosomal which include both reciprocal and non-reciprocal translocations (Fig. 1A, B). Translocations, particularly unbalanced (e.g., with deletion) have been long known to contribute to tumorigenesis [2].
3.1. Distribution of rearrangement endpoints in the human genome
For every chromosomal re-arrangement, COSMIC reports genomic coordinates of two breakpoints labeled as “from” and “to”. For example, if the red segment was deleted in Fig. 1A, the 1st (from) and 2nd (to) breakpoint are given as the coordinates flanking the deletion. The deletion could have occurred because of two breaks that were fused without the intervening segment or one break that was bidirectionally resected as in SSA. Some COSMIC data is from whole genome sequnces while other data from microarray analysis which can estimate the position of the break based on the resolution of the probe. Becasue in this study we are interested in creating a general map of rearangemnets, we refer to these “breakpoints” as “rearrangements endpoints”.
We graphed the first and second rearrangement endpoint genomic coordinates of all cancer tissues with the goal to understand if their distribution is random (Fig. 1C). The histogram distribution of these endpoints clearly identifies peaks with a higher frequency of rearrangements. We immediately note that some chromosomes are characterized by more rearrangements than others suggesting that chromosomal aberrations in the genome are not random. Some of these peaks correlate with previously identified fragile sites [33]. Additionally, except for a few cases, the “from” and “to” endpoints generally correlated with each other in agreement with the observation that most aberrations are intra-chromosomal. Most intrachromosomal deletions are short (1000−10,000 bps average) agreeing with previous analysis [33]. We also find that deletions are also independent of chromosome size or number suggesting that they occur by the same mechanism throughout the genome (Fig. 1D). The distance between the breakpoints of the other intra-chromosomal aberrations (insertions, inversions and duplications) are further apart (about 100,000 bp average) but also not dependent on the chromosome number (Fig. 1E). These aberrations do not eliminate a region of the genome as deletions do and there may be selective pressure against large deletions.
Although most deletions are short, large deletions including deletions of entire chromosome arms do occur (Supplemental Fig. S2). Considering that such large deletions will probably remove essential genes it is likely that they cause hemizygosity and not homozygosity. However, the COSMIC file does not have information on ploidy, and we could not establish zygosity. We again note that the large deletions are not specific to a region of the genome but are found on every chromosome. No one chromosome appears to have a higher density of deletions than any other chromosome, nor does there appear to be a region of any chromosome that is cold to large deletions (Supplementary Fig. S3). Therefore, we conclude that no genomic region prohibits large deletions. Previous studies have also shown that deletions in many cancers have at least a bimodal distribution and in many cases multimodal. In some cancers they show a peak at 1 kb and another at 100Kb but often they distribute anywhere from under 1 kb to over 10Mb [33].
We next identified rearrangement endpoint peaks in the human genome (Fig. 2, Supplementary Fig. S3). To do this, we graphed the coordinates of both the first and second break point together (from and to). The labels represent peaks that appear in every chromosome. This graph clearly shows a higher frequency of breaks in some parts of the genome (e.g., chromosomes 8, 11 and 17). Some of the peaks have already been previously identified [9]. For example, it is known that cancers are in general characterized by chr17p and q CNVs. The tumor suppressor TP53 as well as other cell cycle regulators which are often inactivated in cancers are found on chromosome 17. Chr8p CNVs have also been identified in colorectal and liver cancers. Remarkably, we also found peaks on chromosomes 11, 18, 19, 20 and 21 that to our knowledge were not previously identified. We discuss these peaks in the next section.
Breaks in repetitive regions are more likely to produce rearrangements because any repeat can be used as a template for repair. To understand whether peaks correlate with repetitive regions, we overlayed four tracks from UC Santa Cruz Genome browser (interrupted repeats, microsatellites, segmental duplications and simple repeats) on top of the chromosome break peaks (Supplementary Fig. S3). Many of the high peaks do occur in areas with a higher frequency of segmental duplications (e.g., 7q11.21, 9q13, 10q11.22, 10q23.2, 12q15, 12q24.33, 14q11.2, 14q32.33, 16q23.1, 17q11.2, 17q12, 21p11.2). Segmental duplications are low copy repeats (LCRs) which can stall replication forks and cause DNA damage [38] but at the same time allow homologous recombination repair [39]. Segmental duplications arise by a form of HR repair known as Non-allelic Homologous Recombination (NAHR) which constitutes recombination events between repeats not between two alleles [40]. NAHR can cause, duplications, inversions, deletions as well as other forms of CNVs. These observations suggest that most of the high peaks result from breaks repaired by homologous recombination. A previous study has found that replication origins correlated with rearrangement endpoints [33] suggesting that these breaks may arise form improperly rescued stalled or collapsed replication forks. Most of the smaller peaks do not appear to correlate with segmental duplications or the other forms of repeats which may suggest that they are repaired by NHEJ and related mechanisms.
3.2. The distributions of genome rearrangement endpoints are specific to each cancer type
We next wanted to understand whether the distribution of chromosome breaks in the human genome correlate with the types of cancer. We graphed each rearrangement endpoint by cancer type (Supplementary Figs. S4, 3). This analysis clearly shows that each cancer has a specific rearrangement endpoint signature. To ensure that these distributions are statistically significant, we used the Kolmorogov-Smirnov test for uniformity. A significant probability value (e.g. p < 0.05) indicates that the peaks are not uniform but cluster in different regions of the genome. Every cancer, except cancers of the upper aerodigestive tract shows clustering. This is also obvious when we generated graphs of combined endpoints to determine genome wide rearrangement endpoint density by cancer type (Fig. 3). Thus, it is reasonable to conclude that chromosomal breaks are dependent on the tissue where they occur.
Fig. 3. Genome wide aberration endpoint density by cancer type.
The distribution of the combined first and second aberration endpoint is separated by cancer type. Breakpoint resolution is graphed at one million base pairs.
3.3. The type of chromosomal aberrations is also specific to each cancer
To further understand whether each cancer type is characterized by breaks on specific chromosome we generated matrices of “from” (first break or endpoint) to “to” (second break or end point) (Supplementary Fig. S5). We then generated 3D graphs that represent first break point (X-axis) and second breakpoint (Z-axis) with the fold increase in incidence displayed on the Y-axis (Fig. 4A). For most cancers this produced diagonal peaks consistent with previous findings that most aberrations are deletions. However, three cancers are characterized by an increase in translocations (bone, urinary tract, upper aerodigestive tract). Fig. 4B summarizes the chromosomes that have the highest incidence of breaks in each cancer. Besides chromosome 17 which was expected to be frequent, many cancers are characterized by a high incidence of breaks on chromosome 19 followed by 20 and 21. Most remarkable is that, for the most part, breaks on the longest chromosomes (1–10) do not appear at high incidence in cancers. The lung is the exception where chromosome 1 is involved in intrachromosomal translocations.
We next mapped the copy number variations present in each cancer with the goal to identify cancer specific signatures (Fig. 4C). We find that each cancer is characterized by a specific combination of intra- and inter-chromosomal aberrations. Cancers of the urinary tract are the only ones that are characterized almost exclusively by inter-chromosomal aberrations (e.g., translocations). To parse out intra-chromosomal aberrations in more detail, we created spider graphs separated by cancer type (Supplementary Fig. S6). The spider graphs show intra-chromosomal aberrations, but this visual representation clearly highlights the fact that each cancer has a unique intrachromosomal aberration. Further, we generated circos graphs to map the inter-chromosomal aberrations in each cancer and also find that inter-chromosomal aberrations (e.g., translocations) show a specific cancer signature (Fig. 5). For example, the lung cancers have a higher incidence of Chromosome 1 translocations as discussed above. Taken together, these data suggest that chromosomal re-arrangements are cancer specific and possibly driven by selective pressure in each cancer type.
3.4. Mutation spectra of tumors discussed in this report
Although certain cell cycle regulators (e.g., TP53) are altered or inactivated in many cancers, tissue specific mutations have been identified [41]. To understand whether the genome re-arrangements observed in the cancers discussed in this report correlate with specific mutation signatures, we queried relevant literature including the papers reporting these studies (Table 1).
Table 1.
Mutation spectra highlights of tumors discussed in this report.
| Tissue | Mutation spectra1 | References |
|---|---|---|
|
| ||
| 2Pediatric adrenocortical carcinoma: mutations TP53, ATRX, CTNNB1. | [42,44,45] | |
| Adrenal gland | 3Other studies: Chr19 amplification (CYP gene family), Chrlp and Chr7p loss: benign to malignant transition. | |
|
Osteosarcoma: mutations PI3K/mTOR, ADAM6, TP53, NBPF10, RB1, Chondrosarcoma: mutations COL2A1, TP53, RB1, Hedgehog pathway, IDH1, PRMT5, IDH2. |
[48–54] | |
| Bone |
4COSU486 osteosarcoma,
chondrosarcoma, other rare subtypes: mutations TP53, SUFU, IDH2 Other studies: Osteosarcoma, mutation Chr6p12.3 amplification (COPS3, CCNE1, CDK4, MYC), TP53, RB1; Chondrosarcoma, IDH1, IDH2 mutations Basal like breast cancer: mutations JAK2, IRAK2, CSMD1, NRK, MAP3K8, PTPRJ, WWTR1, TP53, MYCBP2, SNED1; deletions Chr5 (CTNNA1), Chr8 (NRG1), MECR; various translocations. Breast tumor cell line: translocations t(4;11) (q32;q21) (MRE11 truncation); mutations apoptosis, MAPK signaling, cell adhesion, cytoskeleton organization and cell cycle. Primary breast cancer: mutations TP53, GATA3, PIK3CA, MAP2K4, SMAD4, MLL2, MLL3, NCOR1; amplification ERRBB2, CCND1, MYC, MDM2, ZNF217, ZNF703; BRCA1/BRCA2 germline mutation (loss of 17q21 and 13q12). Primary breast cancer: mutations AKT2, ARID1B, CASP8, CDKN1B, MAP3K1, MAP3K13, NCOR1, SMARCD1 and TBX3 Multifocal breast cancer: mutations TP53, PIK3CA, MAP3K1, PTEN, others. |
|
| Breast |
Breast cancers (various): mutations TP53, PIK3CA, MYC, CCND1, ERRBB2, RB1, GATA3, MAP3K1, MAP2K4, others. COSU385 breast triple negative lobular cancer: mutations PIK3CA, TP53, RB1, PTEN, RP11–245C23.3, others; CNVs 1q trisomy, 4q monosomy, sub-clonal del 7, 13, 22q11, other re-arrangements that include MYC, HER2, BRCA1, BRCA2 COSU652: mutations PIK3CA, TP53, PTEN, RB1, ERRB2, RP11–245C23.3 others. COSU668: mutations PIK3CA, RP11–245C23.3, TP53, others Triple negative breast cancer: amplified 4q28.3, 2p, 3q24, 1q21.2, 10p, 12p11.1, 8q 20p11.22–20p11.21, 21q22.13, 6p22.1; deleted 1p36.23, 4q21.1 and 5q. Primary breast tumors: amplified 17q11.2–12 (ERBB2). Breast cancers (various): Chr17 centromere amplification (ERBB2), HER2 amplification. Medulloblastoma: mutations KDM6A and other KDM members, inactivation of members that regulate the H3K27me3 histone mark, WNT signaling mutations, ZMYM3, CTNNB1; Deletion 9q34.14 (DDX31, AK8, TSC1); amplification OTX2. |
[55–63] |
| Central Nervous System |
Pediatric high-grade gliomas: mutations in genes that regulate H3K27me3, histone H3 genes H3F3A (H3.3) and HIST1H3B (H3.1), ATRX, TP53, NTRK1/2, RAS-PI3K, ACVR1. COSU379 medulloblastoma SMO, BRAF, PIK3CA, TP53, RP11–286H14.8, FGFR1, PTEN, RP11–350N15.4, H3K27me3 dysregulation; germline TP53, PTCH1, APC, CREBBP |
[64–71] |
| Endometrium | [72–76] | |
| COSU677 (Uterine cancer, carcinosarcoma): mutations TP53, PTEN, KRAS, PIK3CA, RP11–245C23.3, various other cell adhesion and cell-cell interaction genes, signal transduction genes. | ||
| Type II EC and serous ovarian cancer: amplified 19q12 in.(CCNE1, URI) | TCGA and ICGC study: CSMD3, TP53, APOB, others. | |
| Uterine carcinosarcomas: amplified 19q13 in.(TGFBl). | ||
| Epithelial ovarian cancer: amplified 19p13.11 in. (MERIT40, ANKLE1) | ||
| Hematopoietic and Lymphoid |
Adult T-cell leukemia/lymphoma (ATL): HTLV-1 integration; mutations CDKN2A, ATXN1, NF-kB signaling. |
[77–83] |
|
Early T-cell precursor acute lymphoblastic leukemia (ETP ALL): RAS signaling mutations (BRAF, JAK1/3, KRAS, NRAS FLT3, IGFR1), chromatin modification and remodeling (EZH2, EED, SUZ12, SETD2, EP300), other mutations specific to hematopoetic and lymphoid cell development, some novel mutations. Pediatric B-acute lymphoblastic leukemia: RAS signaling mutations (KRAS, NRAS, PTPN11, FLT3), epigenetic regulators (MLL2), cell cycle regulation (CDKN2A/B deletion, TP53 mutations), other pathways. Chronic lymphocytic leukemia (CLL): mutations NOTCH1, MYD88, XPO1, KLHL6; deletions of 6q14q22, 13q14. COSU340 (Chronic lymphocytic leukemia): mutations TP53, BRAF, KRAS, others COSU440 (Burkitt lymphomas): translocations t(8;14)(q24;q32), t(2;8), t (8;22); mutations ID3, SMARCA4, DDX3X, TP53, BRAF, MYC, RHOA, FBXO11, others. Other ATL studies: TP53, FAS, CCR4, NOTCH1, ZEB1, and CDKN2A. |
||
| Large Intestine | Colorectal adenocarcinoma: translocations VTI1A-TCFL2 (TCF4) fusion, chr8–20, chr5–11; deletions EGFR, PTEN; rearrangements MACROD2, A2BP1, FHIT, IMMP2L; mutations TP53, KRAS, APC. | [84] |
| Liver |
Hepatocellular carcinoma: multiple tumors with hepatitis B (HBV) and C (HCV) viral infections, two multicentric tumors sets: mutations TP53, CTNNB1, ATM, ERRFI1, WWP1, ZIC3, chromatin regulators ARID1A, ARID1B, ARID2, MLL, MLL3, others; HBV integration in TERT region COSU322 (Viral associated hepatocellular carcinoma (Japanese cohort)): common mutations likely pathogenic TP53, RB1, PIK3CA, KRAS, NRAS, EGFR2, others including CSMD1/3, CTNNA2/3, LSAMP, CTNAP2, PCDH15, ALK, RBFOX1, chromatin remodeler HDAC9. |
[85] |
| Lung |
Small cell lung cancer (cell line, smoking): mutations TP53, RB1, MLL2; rearrangements 1p32–36, 4q25–28, 3q, 5q; fusions, PVT1-CHD7 CREBBP2-BTBD12. Small cell lung cancer (surgical resection): mutations PTEN, PIK3CA, TP53, RB1, SLIT2, CREBBP, EP300, others; deletions 3p,13q (RB1 region), 17p (TP53 region); amplifications 3q (SOX2 region), 8p12 (FGFR1 region), 19q12 (CCNE1 region) Primary and metastatic lung adenocarcinomas (NSCLC): various alterations of IQGAP3; overexpression APOBEC1, APOBEC3B, APOBEC3C, APOBEC3F; mutation BRAF, TP53, EGFR, LRP1B, KRAS, PTPRD, STK11, SMAD2, PIK3CA, BRAF, FLT1, RHPN2, GLI3, MRC2; rearrangements 1q, 3q, 5p, 7p/q, 8q, 14q, 16p, 17q, 20q; deletions 3p, 4q, 6q, 8p, 9p, 12q, 13q, 15q, 17p, 18q. |
[86–88] |
| Pancreas |
COSU328, 382 (Ductal adenocarcinoma): mutations KRAS, TP53, SMAD4, CDKN2A, PIK3CA, SMAD4. COSU586 (endocrine neoplasms): MEN1, BRAF, CDKN2A. |
[89] |
| Prostate |
COSU661 (rare pancreatic tumors): TP53, PTEN. Primary prostate cancer: mutations SPTA1, SPOP, TMPRSS2, chromatin modifiers CHD1, CHD5, HDAC9; re-arrangements involving CHD1, PTEN, MAGI2, CSMD3, ZNF407, TP53, MAP2K4, ABL1; fusion TMPRSS2-ERG. Aggressive prostate tumors: BRCA2, ERG, TMPRSS2, KMT2C, TFDP1, VPS13B, FOXA1, PARK2, PTEN, SPOP, TP53. COSU534 (early onset prostate cancer): TP53, PTEN, BRAF, SLC2A2. COSU537, 538, 675 (prostate adenocarcinoma): PIK3CA, RP11245C23.3, TP53, BCL10, AR, PTEN, CTD-2350C19.1, BRAF, XPC, SARM1, SLC46A1, CTD-2350C19.2, KRAS. COLO-829 cell line (metastatic malignant melanoma): mutations, SPDEF, MMP28, UVRAG, BRAF; deletion, PTEN; rearrangements MAGI2, FHIT, WWOX, FRA3B, FRA16D; amplification chr3 (RARB, TOP2B, NGLY1, OXSM), chr15 (MKRN3, NDN) |
[90,91] |
| Skin |
Metastatic melanoma: mutation, BRAF, NRAS, KIT; rearrangements, FHIT, MACROD2, CSMD1, PTEN, MAGI2, A2BP1, ETV1, PREX2. COSU656 (skin adenocarcinoma): mutations BRAF, NRAS, KRAS, MARCh9, TSPAN31 CDK4, CSDE1. |
[92,93] |
| Soft Tissue | Rhabdomyosarcoma: mutations, HRAS, KRAS, NRAS, FGFR4, PIK3CA, NF1, FBXW7, BCOR, TP53, CTNNB1, BUB1B, FOXM1, CCND1, CCND2, BCOR, PTPN11, ATM, ZNF350, TRPC4AP, FOXO1, ARID1A; translocations, t(2;13) (PAX3-FOXO1), t(1:13) (PAX7-FOXO1), chr2q (PAX3-INO80D); other structural variations affecting MIR17HG, CNR1, CDKN2A, ERBB4, RPTOR, FRS2, CACNA1A, NRG1, FOXP2; CNVs: LOH 11p15.5 (IGF2); amplification 12q13-q14 (CDK4), 12q5 (FRS2, MDM2), 2p24 (MYCN), PAX7-FOXO1, 13q21–32 (MIR17 G); homozygous deletion, CDKN2A. | [94] |
| Upper Aerodigestive Tract | Head and neck squamous cell carcinoma: mutations, PTEN, PIK3A, TP53, CDKN2A, HRAS, NOTCH1, NOTCH2, NOTCH3, IRF6, TP63, SYNE1, SYNE2, RIMS2, PCLO, CASP8, DDX3X, PRDM9, EZH2; deletion, CDKN2A, NOTCH3; amplification, CCND1, MYC, EGFR, ERBB2, CCNE1; | [95] |
| Urinary tract |
Bladder cancers of various stages: mutations, CDKN1A, TP53, FGFR3, ARID1A, KDM6A, STAG2, B3GNTN9, FAT1, MLL2/3, PIK3CA, others; amplification 1q, 3q, 8q, chr11q12–3 (CCND1), chr4p16.3 (FGFR3), chr12 (MDM2, NUP107, CPM, CPSF6, LYZ, YEATS4, FRS2); deletion chr9, chr9p21.3 (CDKN2A), chr4p16.3 (FGFR3), PPARGC1A, RFX3, FAT1; chromothripsis 3p, 5q, 6p, X; |
[96] |
Highlited are some of the most likely mutated genes in the indicated cancers. Although other mutations exist, these are mutations specific to these cancers reported in the referenced articles. Please see referenced studies for more in-depth analysis.
These references (bold) include the main studies used for the graphs and analysis in this paper. Please see Supplementary Table S2 for the list of all studies used for the analysis in this paper.
These references (italicized) represent other studies characterizing similar cancers.
For COSU samples only the most frequent pathogenic mutations are listed as reported on https://nam11.safelinks.protection.outlook.com/?url=https%3A%2F%2Fdcc.icgc.org%2F&data=04%7C01%7Cp.punniyaseelan%40elsevier.com%7C08d67d677a034b2df78b08d9d126edcd%7C9274ee3f94254109a27f9fb15c10675d%7C0%7C0%7C637770788370273669%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000&sdata=jqYHDYZK9WZTbVU4V5tV4B3X55bKutYumGyX3yoYxVI%3D&reserved=0. Please see this resource for further information.
3.4.1. Adrenal gland
The adrenal gland re-arrangements from a study on pediatric adrenocortical carcinomas [42] were characterized by TP53 mutations with more than half harboring a previously reported germline mutant (TP53-R337H) discovered in a Southern Brazilian population [43] and known to increase the incidence of these cancers. Certain samples had co-occurring TP53 and ATRX mutations which were associated with increased chromosomal re-arrangements. Other studies have reported amplification of chromosome 19 regions which include the CYP gene family [44] as well as other chromosome gains (chromosomes 5, 12, 4) and losses (chromosomes 1, 2, 11, 17) [45]. The CYP family genes have been implicated in modulating the activity of chemotherapeutic agents [46] and our analyses also shows that chromosome 19 aberrations are high in adrenal gland cancers (Fig. 4A). Losses of chromones 1p and 17p correlate with benign to malignant transition [45,47]. The pediatric adrenocortical carcinomas were also characterized by deletion of 4q34 which encompasses LINCO00290 and amplification of chromosome 9q, while translocations causing fused genes were not frequent [42]. Our analysis showed that most re-arrangements endpoints in pediatric adrenocortical carcinoma occur at chr19q, a region characterized by segmental duplications [97,98]. We identified the CDK2 cyclin CCNE1 cyclin in this region.
3.4.2. Bone
The two bone studies catalogued on COSMIC are for osteosarcoma [48] and chondrosarcoma [49]. Common heterozygous TP53 germline mutations were found in osteosarcoma which were often accompanied by loss of heterozygosity of the WT allele. TP53, RB1, and CDKN2A/B gene deletions were also observed. Osteosarcomas were also characterized by amplifications of CCNE1 and Chr. 6p12.3, which the authors indicate that have been reported in previous studies [50–53]. But perhaps the most important finding from the osteosarcoma study is alterations of the PI3K/mTOR pathway which the authors showed that is essential for osteosarcoma proliferation.
Chondrosarcomas were characterized by COL2A1 (Type II collagen alpha chain) mutations as well as large scale re-arrangements with breakpoints within the COL2A1 coding sequence on chromosome 12 [49]. TP53, RB1 and hedgehog pathway alterations were also identified. Additionally, IDH1 and IDH2 mutations were also seen though this was not surprising because it had been previously reported [54]. IDH1 R132H mutations are prevalent in astrocytomas which correlates with better patient survival [99]. IDH1 functions as a dimer but heterozygous IDH1-R132H/IDH1-WT converts alpha-ketoglutarate into D-2-hydroxyglutarate which is a known oncometabolite [100]. Deletion of 19q and 1p co-occurs with IDH1 mutations in gliomas and it has been proposed that IDH1 mutations are a prerequisite for these translocations [100]. We also identify an increase breakpoint frequency within chromosome 19 in bone cancers (Fig. 4B). Remarkably, all of the chondrosarcoma IDH1 mutations in the Tarpey et al. study substitute arginine for a different amino acid than histidine at residue 132, most often cysteine, though previous studies have identified the R132H mutation. We have shown that in astrocytomas IDH1 is often commutated with TP53, ATRX and NOTCH1 but not CHEK2 and PTEN [101] which remarkably mirrors the two bone cancers discussed here: osteosarcoma has PTEN (within the PI3K/mTOR) mutations but not IDH1 while chondrosarcomas have IDH1 mutations but not PTEN. IDH1/2 mutations have been identified in other studies (Table 1) including in the COSU486 study reported on COSMIC highlighting the importance of these genes in bone cancer.
3.4.3. Breast
One of the breast cancer studies reported on COSMIC is a basal like breast cancer mouse xenograft from a patient [55] on neoadjuvant chemotherapy [102]. The most prevalent mutation was a deletion on chromosome 5 that includes CTNNA1 (catenin alpha 1) with roles in cell adhesion. Besides TP53 most mutations were in signal transduction pathways and cell cycle regulation (Table 1). No mutation was reported in DNA damage repair genes. Another study is of the breast tumor cell line HCC1954 [56,103]. HCC1954 is characterized by several structural variations [104] causing MRE11 truncations [105]. Gene network analysis by Galante et al. identified that the higher mutation burden is in the apoptosis, MAPK signaling, cell adhesion, cytoskeleton organization and cell cycle pathways. Two studies analyze primary breast cancers [57,58]. In one of the studies (COSU652) samples had mutations in cell cycle regulation signal transduction pathways (e.g., MAP2K4, PIK3CA, ERBB2) (Table 1) [57]. Mutations in both PIK3CA and the antisense transcript RP11–245C23.3 which overlaps with PIK3CA coding region were common in breast cancers. Mutations in TP53 and TP53 regulatory factors (MDM2) were also observed. A subset of the samples had BRCA1 and BRCA2 germline mutations and were characterized by chromosome 17q21 and 13q12 loss. The other study [58] also identified mutations in signal transduction pathways that regulate the cell cycle. A study of multifocal breast cancer [59] and another on 560 breast cancers [60] also identified mutations in similar genes (Table 1). The latter also identified mutations in RB1 which none of the other report.
A search of the literature also identified that triple negative breast cancers are characterized by significant chromosomal re-arrangements [61]. A region characterized by segmental duplications were identified as the potential culprit for amplification of a chromosome region encompassing ERBB2 (17q11.2–12) in primary breast cancers [62]. Chromosome 17 centromere amplification that includes the ERBB2 was also detected in other breast cancers [106]. In the latter case ERBB2 co-amplification with HER2 was common. We also observe that most rearangement endpoints in our analysis fall on chromosome 17 which is consistent with these studies. An association between ERBB2 amplification and general chromosomal re-rearrangements has been proposed and is reviewed here [107].
3.4.4. Central nervous system (CNS)
COSMIC reports two CNS studies, one from medulloblastoma and one from pediatric high-grade gliomas. Both were characterized by mutations in factors that establish or regulate the histone H3K27me3 mark which is required for transcriptional regulation of certain stem cell genes [108]. Pediatric high-grade gliomas were also characterized by mutations in the histone genes H3F3A and HSIT1H3B [69]. Changes in this chromatin mark is most often associated with altered gene expression but it also affects NHEJ repair through interaction with FANCD2 [109]. The authors also point out that certain germline mutations have a predisposition to medulloblastoma [65–68] but they were rare in this study. The WNT signaling pathway was also altered in medulloblastoma, most often mutations that increase CTNNB1 stability. The WNT pathway controls cell proliferation, cell migration as well as other embryonic development processes and mutations in this pathway are common in medulloblastoma [110]. Additional mutations in high grade gliomas include ATRX, TP53 as well as deregulation of certain signal transduction pathways (Table 1) [69]. Some of the samples reported on COSMIC are from the COSU379 study (pediatric brain cancers) which are also characterized by mutation in histone genes or H3K27me3 dysregulation [70,71].
3.4.5. Endometrium
The most common mutations in these cancers are in cell adhesion and cell-cell interaction genes (Table 1). There were no apparent mutations in chromatin remodelers, DNA damage repair, DNA replication or transcription. Remarkably, there were reported mutations in CSMD1 [111] and CSMD3 [112], two genes that are involved in neuronal development. A recent study also identified CSMD3 mutations in ovarian cancers [72] and showed that CSMD3 mutations are associated with poor prognosis. This study also identified several other mutated genes including TP53 and APOB. Several other studies report Chr19 CNVs [73–76] and we also find that most breakpoints are on chromosome 19 (Fig. 4).
3.4.6. Hematopoietic and lymphoid
One study is of adult T-cell leukemia/ lymphoma (ATL) [77]. Human T-cell leukemia virus type-1 (HTLV-1) infection is a hallmark of this cancer [113] and the authors point out that mutations in TP53, FAS, CCR4, NOTCH1, ZEB1, and CDKN2A have been previously identified [78–80]. The current study identifies mutations in components of the NF-kB pathway and the author postulate that mutations generally allow cells to avoid host immune surveillance. A second study of early T-cell precursor acute lymphoblastic leukemia (ETP ALL) identified mutations in RAS signaling pathway and chromatin remodeling/modification [81]. Of note are mutations in EZH2 (H3K27me3) and SETD2 (H3K36me3). A third study is of pediatric B-acute lymphoblastic leukemia also identified mutations in the RAS signaling pathway, epigenetic modification genes (MLL2) cell cycle regulation genes (CDKN2A/B and TP53) as well as other pathways [82]. In this study, the authors followed cells from diagnosis to relapse with mutations in NT5C2 found specifically in relapsed patients. NT5C2, a gene required for dephosphorylation of various 6-hydroxypurine nucleotide monophosphosphates prior to cellular export [114,115] but also inactivates nucleoside analog drugs. NT5C2 activating mutations have been observed in chemotherapy resistant cells [116]. A fourth study on chronic lymphocytic leukemias (CLL) also identified mutations in NOTCH1, and other immune system specific genes [83]. NOTCH1 was also identified in the COSU340 CLL study which is also reported on COSMIC [117]. Finally, some samples are from the COSU440 study on Burkitt Lymphomas [118] which are characterized by chromosome 8 translocations all of which happen within the MYC oncogene locus (8q24) and cause MYC dysregulation [119]. Notably, they identified ID3, a gene involved in regulation of cell cycle progression as well as other processes. One function of ID3 is to negatively regulate E-box sequence transcription factor TCF3 which is required for Burkitt Lymphoma proliferation and viability [120]. Consequently, ID3 and TCF3 mutations are common in these cancers.
3.4.7. Large intestine
Primary colorectal adenocarcinoma tumors reported on COSMIC are characterized by VT11A-TCF4 fusion [84]. TCF4 is a helix-loop-helix transcription factor related to TCF3. It has roles in nervous system development and with certain dominant mutations causing the autism related Pitt Hopkins syndrome [121]. It also interacts with CTNBB1 (beta catenin) and regulates transcription of genes involved in intestinal epithelial cells growth [122]. VT11A-TCF4 fusions were also found in a colorectal cell line [84]. Although, this fusion is not dominant negative, the authors speculate that it could allow TCF4 to activate transcription of its targets in the absence of CTNBB1 considering that this fusion is found in the background of APC mutations. APC is a known beta catenin suppressor. Other mutations in cell cycle regulators (TP53, KRAS, PTEN) were also identified in the large intestine cancers. Fusions between chromosomes 8 and 20 and chromosomes 5 and 11 were also identified. Our analysis also found that aberrations endpoints were most abundant on chromosome 20.
3.4.8. Liver
Hepatocellular carcinoma with and without hepatitis B (HBV) and C (HCV) viral infections were analyzed [85]. Mutations in cell cycle regulators (TP53, ATM) as well as chromatin regulators (ARID1A, ARID1B, ARID2, MLL, MLL3) were detected in most tumors with no statistical differences between HBV and HCV status. Viral integration in the TERT region was also detected in HBV tumors. COSU322 was the second study from viral infected Japanese donors. Previously reported mutations in cell cycle regulators (e.g., TP53, RB1, PIK3CA, KRAS, NRAS, EGFR2) [41] were detected. Mutations in CSMD1 and CSMD3 were also reported in these studies as well as mutations in cadherins and other cell adhesion and migration proteins. A mutation in the chromatin remodeler HDAC9 was also identified.
3.4.9. Lung
A small cell lung cancer (SCLC) cell line from a smoking patient [123] had characteristic SCLC gene mutations (TP53, RB1, others) which the authors point out that they have been previously reported in the literature [124–127]. Several unbalanced intrachromosomal translocations were identified as well as intra-chromosomal inversions. One translocation fuses CREBBP with BTBD12 (SLX4) a Holliday junction resolvase involved in DSB repair [128,129]. Elevated levels of CHD7 transcripts were detected due to a PVT1-CHD7 fusion and amplification of the PVT1 locus. CHD1 binds methylated H3K4 at enhancer and promoter regions [86]. Two cases had both MYC1 and CHD1 high levels of gene expression suggesting that CHD1 amplification may drive MYC1 high expression levels. These findings are mirrored by a second SCLC study of surgically resected specimens (Table 1) [87]. The authors also showed that mutations in the chromatin remodelers CREBBP and EP300 reduced H3K18 acetylation, a mark that is involved in transcription elongation [130]. A third study is of non-small-cell lung cancer (NSCLC) [88]. Mutations in several known genes (TP53, PIK3CA, etc.) were identified as well as some novel genes (GLI3, MRC2). Several APOBEC genes were upregulated which correlate with C > A mutations. Several known chromosomal aberrations were also identified (e.g., 3q, 17q) as well as some structural aberrations involving cytoskeleton regulation genes (IQGAP3 and others) and cell cycle regulation genes (CHEK2, FGFR2, etc.).
3.4.10. Ovary
The reported ovarian cancer samples are from the COSU585 study (serous cystadenocarcinoma). These samples are characterized by TP53 and KRAS mutations. Additionally, mutations in certain mitochondrial genes involved in sugar metabolism and respiration (e.g., MT-CYB, MT-ND2) or mitochondrial tRNAs (e.g., MT-TI, MT-TW) were also identified. A connection between mitochondrial oxidative phosphorylation, BRCA1 and homologous recombination has been previously made in several cancers including ovarian cystadenocarcinoma [131]. Changes in reactive oxygen species affect expression levels of BRCA1 and biases repair through NHEJ rather than HR. Unfortunately, gene expression levels for these samples are not available and we could not verify its status. However, we do find that these cancers are characterized by deletions and duplications which may suggest BRCA1 independent pathways.
3.4.11. Pancreas
The pancreatic data is from five studies (COSU328, COSU382, COSU586, COSU650 and COSU661). Ductal adenocarcinomas are characterized by TP53, KRAS, SMAD4, CDKN2A, and PIK3CA mutations [89]. Some rare pancreatic tumors (COSU661) have PTEN and TP53 mutations. MEN1 is a characteristic mutation of endocrine neoplasms [132]. Some MEN1 mutations give rise to certain parathyroid syndromes but we have shown that these mutations do not predispose patients to cancer [133].
3.4.12. Prostate
Primary cancers are characterized by mutations in chromatin remodeling genes (CHD1, CHD5, HDAC9), and chromosomal re-arrangements involving several cell cycle regulators (PTEN, MAGI2, TP53, MAP2K4, CSMD3, CADM2 TMPRSS2-ERG fusion etc.) [90]. PTEN and ERG dysregulation has been shown to occur in aggressive prostate tumors [134,135]. Re-arrangements occurred primarily in open chromatin regions, but the authors do conclude that translocations affect mainly regions and genes that promote tumor development. Aggressive prostate tumors were characterized by biallelic inactivation of BRCA2 [91], either somatic or with one inherited germline mutation in addition to the known culprits. Similar mutations were found in the four COSU cohorts including some prostate specific genes (Table 1).
3.4.13. Skin
A cell line (COLO-829) from a metastatic malignant melanoma shows mutations characteristic of UV damage and transcription coupled repair [93]. Mutations in certain cell cycle regulators, cell adhesion and proliferation, transcription regulators as well as others were also identified (Table 1). BRAFV600E, the signature mutation in melanoma [136, 137], was identified in these cancers. Sequencing of 25 metastatic melanomas identified similar mutations [92]. Chromosomal re-arrangements around and within PREX2 locus as well as amplification of this gene was detected. PREX2 is a PTEN inhibitor and PIK3CA pathway activator [138] suggesting that melanoma cells rely on the activity of the PIK3CA.The COSU656 ICGC study also did not find any predicted mutation in PIK3CA.
3.4.14. Soft tissue
Rhabdomyosarcoma is a mesenchymal cell cancer in skeletal muscle [139]. Mutations were found in various cell cycle regulators and signal transduction pathways (e.g., TP53, NRAS, KRAS, PIK3CA) [94] (Table 1). Several chromosomal re-arrangements involving other cell cycle regulates (e.g., CDKN2A, MDM2) were also identified. Novel mutations were identified in FBXW7, a E3 ubiquitin ligase [140], and BCOR, a histone deacetylase interactor and transcriptional repressor [141]. PAX7-FOXO1 and PAX3-FOXO1 fusions characteristics of these cancers [142,143]. Both the PAX genes and FOXO1 are transcription factors, and these fusions is believed to allow transcriptional regulation of different targets. COSMIC also reports certain cases from the Fujimoto et al. liver study above [85] as fibrous tissue of uncertain origin. Fujimoto et al. found that mutations in chromatin regulators are associated with liver fibrosis.
3.4.15. Upper aerodigestive tract
Squamous cell carcinomas are characterized by mutations in pathways that regulate squamous cell differentiation which includes genes in the NOTCH pathway (NOTCH1, NOTCH2, NOTCH3) [95]. Most patients characterized in this study had a history of alcohol and tabaco use. Previously identified mutations in cell cycle regulators such as CDKN2A deletion and EGFR and TP53 mutations were also identified here [144]. Certain mutations in apoptotic genes (CASP8, DDX3X) and chromatin remodelers (PRMDM9, EZH2) were also detected in a subset of the samples.
3.4.16. Urinary tract
CDKN1A, FAT1, and TP53 mutations as well as MDM2 amplification have been detected in bladder cancers (Table 1) [96]. Other driver mutations (e.g., PIK3CA, CDKN2A, MLL2/3) were also prevalent. Chromothripsis of 3p, 5q, 6p and X were detected. Our analysis also identified chromosomes 4–21 and 2–21 translocations (Fig. 4A, B).
4. Discussion
4.1. Intra vs. inter-chromosomal re-arrangements
Our analysis of the COSMIC data shows that about 75 % of re-arrangements are intra-chromosomal while only 25 % are inter-chromosomal (e.g., translocations), which has also been previously shown by other studies[33]. This is not unexpected because chromosomal translocations are influenced by the organization of the genome in the nucleus [145–148]. Therefore, a break is much more likely to be repaired by ligation to another proximal break than to a distant break. However, this observation is not universal as break mobility is somewhat influenced by the type of damage [149,150], whether the break is in an actively transcribed region [151], the type of repair [152,153], and the cell cycle stage [154,155]. Remarkably, this does not appear to be true in yeast [156] suggesting that in yeast repair is determined only by the type of the repair or the availability of homologous sequence.
It is estimated that human cells encounter 25 DSBs per cell per day [157] though cancer cells that have lost cell cycle regulators or DNA damage repair genes experience many more breaks sometimes in catastrophic events such as chromotripsis or break-fusion-bridge cycles. For a deletion to occur 75 % of the breaks must be within ~10,000bps of each other (Fig. 1D) or within ~100,000bps of each other for other intrachromosomal aberrations (Fig. 1E) assuming no break processing (e.g., resection). Considering that the genome is over 3.1 × 109bps this is unlikely in quiescent cells but much more likely in cancer cells. Thus, deletions would be predicted to occur at higher level in cancer cells and previous studies have shown that NHEJ and related pathways are the primary repair mechanisms because there is little or no homology at the junctions ([33] and references therein). Nevertheless, break repair must go through a form of purifying selection and what we see when we analyze cancer genomes are only those breaks that could be repaired and for which repair conferred an advantage to the cancer cell. That selection occurs is almost certain and is revealed by certain persistent signatures such as deletion of key cell cycle regulators (CDKN2A, TP53, PTEN, etc.) [4–9] or translocations such as the Philadelphia chromosome translocation t(9;22)(q34;q11) in blood cancers [15,158] which activates the ABL1 tyrosine kinase [17] and confers a proliferative advantage to the cancer cells. However, even this translocation is facilitated by the nuclear arrangements of the chromosomes [159] highlighting the importance of proximity in break repair.
4.2. Repair pathway choice and cell cycle stage also determine the type of chromosomal aberrations
In humans most breaks occurring in G1 are repaired by NHEJ (also known as canonical NHEJ, c-NHEJ) and related pathways (backup NHEJ, B-NHEJ; microhomology-mediated end joining, MMEJ) [160] while those occurring in S-phase or G2 are generally repaired by HR [161]. However, the temporal separation of the two repair mechanisms is not quite that clear cut and competition and/or cooperation between the two general mechanisms occurs [162]. Whether a break is repaired by HR or NHEJ may affect the type of chromosomal aberrations that can be produced. Unlike NHEJ, HR needs a homologous region and the search for homology necessitates exploration of a larger nuclear space.
C-NHEJ appears to be the primary mechanism by which re-arrangements occur in human cells [163]. This is supported by the fact that aberration junctions are characterized by small levels of homology [104] and that microhomology mediated end joining is generally dispensable [163]. This suggests that at least two breaks must occur at any given time to produce any re-arrangement.
HR is generally error-free. Because it operates in S-phase and G2, break repair occurs off the sister chromatid which is identical. However, if repair occurs using a homologous region that is not the sister, it could produce re-arrangements. Single strand annealing (SSA) is one backup HR pathway that facilitates repair of breaks occurring between direct repeats [37]. In the case of SSA, repair is found on the same chromosome. More importantly, SSA does not require two breaks. Rather a break is bidirectionally resected until homology is found on either side, then the two homologous regions are annealed to each other [164]. Remarkably, certain tumor suppressors including BRCA1 and MLL sit between repetitive elements [165–167]. Break induced replication (BIR), the major HR pathway for rescue of stalled or collapsed replication forks could also produce non-reciprocal translocations [168]. Our observation that some of the high peaks are characterized by segmental duplications (Supplementary Fig. S3) suggests that suggests that HR, perhaps in the process of rescuing stalled or collapsed forks, causes some of the higher frequency peaks.
Finally, the DNA damage checkpoint also appears to play a major role in whether re-arrangements occur. ATM is the major signal transduction kinase that activates the DNA damage checkpoint in response to DSBs [169]. ATM mutations causes illegitimate DSB repair that produces re-arrangements [170–172]. Further, genetic alterations of p21 which controls the G1/S checkpoint in the p53 mutant background may bias repair towards SSA and BIR at the expense of more error-free HR repair [173,174].
4.3. Reccurent re-arrangements and cancer specific signatures
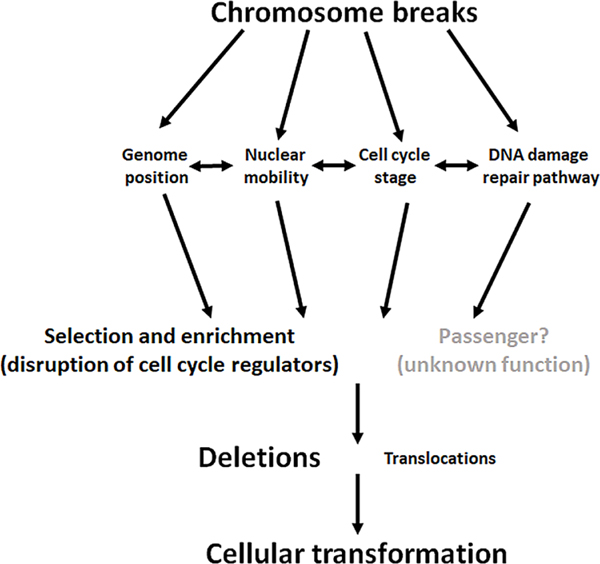
A quick overview of the genes mutated in all these cancers reveals many DNA damage repair and chromatin structure function genes (Table 1). Therefore, it is reasonable to conclude that failure to properly repair chromosome breaks causes re-arrangements. Breaks may occur randomly through the genome and failure to repair properly may be dependent on inactivation of processes involved in DNA damage repair, the cell cycle stage, restrictions or relaxation of nuclear mobility and genome position (Fig. 6). Because the human genome is mainly “junk” DNA many of these re-arrangements may not affect any genes. Alternatively, they may only affect genes that do not contribute to transformation (e.g. same type of genes that acquire passanger mutations). Thus, these CNVs may behave as “passenger” analogous to the many passenger mutations found in all cancers. Because genome position and nuclear mobility restricts break repair, a majority of CNVs are deletions. However, CNVs that occur in key cell cycle regulators are selected for because they confer an advantage to the cancer cell. Indeed, although certain recurrent mutations do occur in most cancers (e.g., TP53, PTEN, CDKN2a), the mutation spectrum of each cancer is unique (Table 1) [41]. But are these recurrent re-arrangements "drivers" analogous to driver genes. The literature certainly refers to them as drivers, particularly if they inactivate key cell cycle regulators [28].
Fig. 6. Model for chromosomal rearrangements in human cancers.
In this model we propose that chromosome break repair is influenced by genome position, nuclear mobility, cell cycle stage and DNA damage repair pathway choice (e.g., HR vs, NHEJ). Those re-arrangements that disrupt cell cycle regulators and confer an advantage to cellular transformation and cancer progression are selected and enriched for whereas others that disrupt essential genes would kill the cells and are eliminated from cancer cell populations. Some re-arrangements that occur in “junk DNA” or affect neither cell cycle regulators nor essential genes are carried through cellular transformation because they may have little effect on cancer progression. These may be “passenger” rearrangements analogous to passenger mutations. Because of restrictions on chromosome mobility within the nucleus, most re-arrangements are deletions.
There remains a question as to whether “passenger” deletions contribute to cellular transformation. Some deletions are known to cause fusion genes (Table 1) [175] which directly implicates them in carcinogenesis. Others have no known function. There is also a question of ploidy. The Knudson hypothesis suggests that tumor suppressors require two hits for inactivation [176]. However, some evidence suggests that haploinsufficiency [177,178] of even “established” tumor suppressors [179,180] is enough to drive cellular transformation, and Knudson himself refined his initial hypothesis [181]. For example, p27Kip1 heterozygous mutations are sufficient to contribute to cellular transformation [179]. A recent elegant review further discusses the role of deletions in cancers [9].
4.4. Conclusion
In this report we show that chromosomal breaks have distinct cancer specific patterns. These breaks also produce distinct cancer specific CNVs. Thus, most chromosomal re-arrangements seem to occur during the evolution of the cancer genome and are specific to each tissue.
Supplementary Material
Acknowledgement
We thank James and Ellen Bazzoli for their generous sponsorship of our laboratory space.
Funding
This project was funded in part by an NIH NCI grant (R03CA252498). Other funding from The Ohio State University James Comprehensive Cancer Center.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.mrfmmm.2021.111773.
References
- [1].Yi K, Ju YS, Patterns and mechanisms of structural variations in human cancer, Exp. Mol. Med 50 (8) (2018) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang WJ, Li LY, Cui JW, Chromosome structural variation in tumorigenesis: mechanisms of formation and carcinogenesis, Epigenetics Chromatin 13 (1) (2020) 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shlien A, Malkin D, Copy number variations and cancer, Genome Med. 1 (6) (2009) 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hamid A, Petreaca B, Petreaca R, Frequent homozygous deletions of the CDKN2A locus in somatic cancer tissues, Mutat. Res 815 (2019) 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Raschke S, Balz V, Efferth T, Schulz WA, Florl AR, Homozygous deletions of CDKN2A caused by alternative mechanisms in various human cancer cell lines, Genes Chromosomes Cancer 42 (1) (2005) 58–67. [DOI] [PubMed] [Google Scholar]
- [6].Lee B, Yoon K, Lee S, Kang JM, Kim J, Shim SH, Kim HM, Song S, Naka K, Kim AK, et al. , Homozygous deletions at 3p22, 5p14, 6q15, and 9p21 result in aberrant expression of tumor suppressor genes in gastric cancer, Genes Chromosomes Cancer 54 (3) (2015) 142–155. [DOI] [PubMed] [Google Scholar]
- [7].Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. , Signatures of mutation and selection in the cancer genome, Nature 463 (7283) (2010) 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cheng J, Demeulemeester J, Wedge DC, Vollan HKM, Pitt JJ, Russnes HG, Pandey BP, Nilsen G, Nord S, Bignell GR, et al. , Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors, Nat. Commun 8 (1) (2017) 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen M, Yang Y, Liu Y, Chen C, The role of chromosome deletions in human cancers, Adv. Exp. Med. Biol 1044 (2018) 135–148. [DOI] [PubMed] [Google Scholar]
- [10].Kubo T, Yamamoto H, Lockwood WW, Valencia I, Soh J, Peyton M, Jida M, Otani H, Fujii T, Ouchida M, et al. , MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors, Int. J. Cancer 124 (8) (2009) 1778–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kakadia S, Yarlagadda N, Awad R, Kundranda M, Niu J, Naraev B, Mina L, Dragovich T, Gimbel M, Mahmoud F, Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma, Oncotargets Ther. 11 (2018) 7095–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rolli V, Gallwitz M, Wossning T, Flemming A, Schamel WWA, Zurn C, Reth M, Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop, Mol. Cell 10 (5) (2002) 1057–1069. [DOI] [PubMed] [Google Scholar]
- [13].Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T, Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer, Cancer Res. 61 (9) (2001) 3550–3555. [PubMed] [Google Scholar]
- [14].Nowell PC, The minute chromosome (Phl) in chronic granulocytic leukemia, Blut 8 (1962) 65–66. [DOI] [PubMed] [Google Scholar]
- [15].Nowell PC, Hungerford DA, Chromosome studies on normal and leukemic human leukocytes, J. Natl. Cancer Inst 25 (1960) 85–109. [PubMed] [Google Scholar]
- [16].Nowell PC, Hungerford DA, Minute chromosome in human chronic granulocytic leukemia, Science 132 (3438) (1960), 1497–1497. [Google Scholar]
- [17].Lugo TG, Pendergast AM, Muller AJ, Witte ON, Tyrosine kinase-activity and transformation potency of Bcr-Abl oncogene products, Science 247 (4946) (1990) 1079–1082. [DOI] [PubMed] [Google Scholar]
- [18].Albertson DG, Collins C, McCormick F, Gray JW, Chromosome aberrations in solid tumors, Nat. Genet 34 (4) (2003) 369–376. [DOI] [PubMed] [Google Scholar]
- [19].Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. , Massive genomic rearrangement acquired in a single catastrophic event during cancer development, Cell 144 (1) (2011) 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zavacka K, Plevova K, Jarosova M, Pospisilova S, Chromothripsis – extensive chromosomal rearrangements and their significance in cancer, Klin. Onkol 32 (2) (2019) 101–108. [DOI] [PubMed] [Google Scholar]
- [21].Leibowitz ML, Zhang CZ, Pellman D, Chromothripsis: a new mechanism for rapid karyotype evolution, Annu. Rev. Genet 49 (2015) 183–211. [DOI] [PubMed] [Google Scholar]
- [22].Umbreit NT, Zhang CZ, Lynch LD, Blaine LJ, Cheng AM, Tourdot R, Sun L, Almubarak HF, Judge K, Mitchell TJ, et al. , Mechanisms generating cancer genome complexity from a single cell division error, Science 368 (6488) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dahiya R, Hu Q, Ly P, Mechanistic origins of diverse genome rearrangements in cancer, Semin. Cell Dev. Biol (2021). S1084–9521(21)00035–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wilson TE, Sunder S, Double-strand breaks in motion: implications for chromosomal rearrangement, Curr. Genet 66 (1) (2020) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mehta A, Haber JE, Sources of DNA double-strand breaks and models of recombinational DNA repair, Cold Spring Harb. Perspect. Biol 6 (9) (2014), a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ensminger M, Lobrich M, One end to rule them all: non-homologous end-joining and homologous recombination at DNA double-strand breaks, Br. J. Radiol 93 (1115) (2020), 20191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kakarougkas A, Jeggo PA, DNA DSB repair pathway choice: an orchestrated handover mechanism, Br. J. Radiol 87 (1035) (2014), 20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, Hornshoj H, Hess JM, Juul RI, Lin Z, et al. , Analyses of non-coding somatic drivers in 2,658 cancer whole genomes, Nature 578 (7793) (2020) 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cortes-Ciriano I, Lee JJ, Xi R, Jain D, Jung YL, Yang L, Gordenin D, Klimczak LJ, Zhang CZ, Pellman DS, et al. , Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing, Nat. Genet 52 (3) (2020) 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Akdemir KC, Le VT, Chandran S, Li Y, Verhaak RG, Beroukhim R, Campbell PJ, Chin L, Dixon JR, Futreal PA, et al. , Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer, Nat. Genet 52 (3) (2020) 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, Santamarina M, Ju YS, Temes J, Garcia-Souto D, et al. , Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition, Nat. Genet 52 (3) (2020) 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Group PTC, Calabrese C, Davidson NR, Demircioglu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, et al. , Genomic basis for RNA alterations in cancer, Nature 578 (7793) (2020) 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, et al. , Patterns of somatic structural variation in human cancer genomes, Nature 578 (7793) (2020) 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. , COSMIC: the catalogue of somatic mutations in cancer, Nucleic Acids Res. 47 (D1) (2019) D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA, Circos: an information aesthetic for comparative genomics, Genome Res. 19 (9) (2009) 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kramara J, Osia B, Malkova A, Break-induced replication: the where, the why, and the how, Trends Genet. 34 (7) (2018) 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Blasiak J, Single-strand annealing in cancer, Int. J. Mol. Sci 22 (4) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].van Binsbergen E, Origins and breakpoint analyses of copy number variations: up close and personal, Cytogenet. Genome Res 135 (3–4) (2011) 271–276. [DOI] [PubMed] [Google Scholar]
- [39].George CM, Alani E, Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA, Crit. Rev. Biochem. Mol. Biol 47 (3) (2012) 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Stankiewicz P, Lupski JR, The genomic basis of disease, mechanisms and assays for genomic disorders, Genome Dyn. 1 (2006) 1–16. [DOI] [PubMed] [Google Scholar]
- [41].Schneider G, Schmidt-Supprian M, Rad R, Saur D, Tissue-specific tumorigenesis: context matters, Nat. Rev. Cancer 17 (4) (2017) 239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pinto EM, Chen X, Easton J, Finkelstein D, Liu Z, Pounds S, Rodriguez-Galindo C, Lund TC, Mardis ER, Wilson RK, et al. , Genomic landscape of paediatric adrenocortical tumours, Nat. Commun 6 (2015) 6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ribeiro RC, Sandrini F, Figueiredo B, Zambetti GP, Michalkiewicz E, Lafferty AR, DeLacerda L, Rabin M, Cadwell C, Sampaio G, et al. , An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma, Proc. Natl. Acad. Sci. U. S. A 98 (16) (2001) 9330–9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rubinstein JC, Brown TC, Goh G, Juhlin CC, Stenman A, Korah R, Carling T, Chromosome 19 amplification correlates with advanced disease in adrenocortical carcinoma, Surgery 159 (1) (2016) 296–301. [DOI] [PubMed] [Google Scholar]
- [45].Sidhu S, Marsh DJ, Theodosopoulos G, Philips J, Bambach CP, Campbell P, Magarey CJ, Russell CF, Schulte KM, Roher HD, et al. , Comparative genomic hybridization analysis of adrenocortical tumors, J. Clin. Endocrinol. Metab 87 (7) (2002) 3467–3474. [DOI] [PubMed] [Google Scholar]
- [46].Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M, Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment, Hum. Genet 127 (1) (2010) 1–17. [DOI] [PubMed] [Google Scholar]
- [47].Stephan EA, Chung TH, Grant CS, Kim S, Von Hoff DD, Trent JM, Demeure MJ, Adrenocortical carcinoma survival rates correlated to genomic copy number variants, Mol. Cancer Ther 7 (2) (2008) 425–431. [DOI] [PubMed] [Google Scholar]
- [48].Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS, et al. , Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma, Proc. Natl. Acad. Sci. U. S. A 111 (51) (2014) E5564–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tarpey PS, Behjati S, Cooke SL, Van Loo P, Wedge DC, Pillay N, Marshall J, O’Meara S, Davies H, Nik-Zainal S, et al. , Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma, Nat. Genet 45 (8) (2013) 923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Henriksen J, Aagesen TH, Maelandsmo GM, Lothe RA, Myklebost O, Forus A, Amplification and overexpression of COPS3 in osteosarcomas potentially target TP53 for proteasome-mediated degradation, Oncogene 22 (34) (2003) 5358–5361. [DOI] [PubMed] [Google Scholar]
- [51].Miller CW, Aslo A, Won A, Tan M, Lampkin B, Koeffler HP, Alterations of the p53, Rb and MDM2 genes in osteosarcoma, J. Cancer Res. Clin. Oncol 122 (9) (1996) 559–565. [DOI] [PubMed] [Google Scholar]
- [52].Wadayama B, Toguchida J, Shimizu T, Ishizaki K, Sasaki MS, Kotoura Y, Yamamuro T, Mutation spectrum of the retinoblastoma gene in osteosarcomas, Cancer Res. 54 (11) (1994) 3042–3048. [PubMed] [Google Scholar]
- [53].Toguchida J, Ishizaki K, Sasaki MS, Ikenaga M, Sugimoto M, Kotoura Y, Yamamuro T, Chromosomal reorganization for the expression of recessive mutation of retinoblastoma susceptibility gene in the development of osteosarcoma, Cancer Res. 48 (14) (1988) 3939–3943. [PubMed] [Google Scholar]
- [54].Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. , IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours, J. Pathol 224 (3) (2011) 334–343. [DOI] [PubMed] [Google Scholar]
- [55].Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. , Genome remodelling in a basal-like breast cancer metastasis and xenograft, Nature 464 (7291) (2010) 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Galante PA, Parmigiani RB, Zhao Q, Caballero OL, de Souza JE, Navarro FC, Gerber AL, Nicolas MF, Salim AC, Silva AP, et al. , Distinct patterns of somatic alterations in a lymphoblastoid and a tumor genome derived from the same individual, Nucleic Acids Res. 39 (14) (2011) 6056–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. , Mutational processes molding the genomes of 21 breast cancers, Cell 149 (5) (2012) 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. , The landscape of cancer genes and mutational processes in breast cancer, Nature 486 (7403) (2012) 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Desmedt C, Fumagalli D, Pietri E, Zoppoli G, Brown D, Nik-Zainal S, Gundem G, Rothe F, Majjaj S, Garuti A, et al. , Uncovering the genomic heterogeneity of multifocal breast cancer, J. Pathol 236 (4) (2015) 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. , Landscape of somatic mutations in 560 breast cancer whole-genome sequences, Nature 534 (7605) (2016) 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pariyar M, Johns A, Thorne RF, Scott RJ, Avery-Kiejda KA, Copy number variation in triple negative breast cancer samples associated with lymph node metastasis, Neoplasia 23 (8) (2021) 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Marotta M, Chen X, Inoshita A, Stephens R, Budd GT, Crowe JP, Lyons J, Kondratova A, Tubbs R, Tanaka H, A common copy-number breakpoint of ERBB2 amplification in breast cancer colocalizes with a complex block of segmental duplications, Breast Cancer Res. 14 (6) (2012) R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. , The life history of 21 breast cancers, Cell 149 (5) (2012) 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. , Novel mutations target distinct subgroups of medulloblastoma, Nature 488 (7409) (2012) 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. , Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms, Science 250 (4985) (1990) 1233–1238. [DOI] [PubMed] [Google Scholar]
- [66].Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B, et al. , The molecular basis of Turcot’s syndrome, N. Engl. J. Med 332 (13) (1995) 839–847. [DOI] [PubMed] [Google Scholar]
- [67].Taylor MD, Mainprize TG, Rutka JT, Becker L, Bayani J, Drake JM, Medulloblastoma in a child with Rubenstein-Taybi Syndrome: case report and review of the literature, Pediatr. Neurosurg 35 (5) (2001) 235–238. [DOI] [PubMed] [Google Scholar]
- [68].Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, et al. , Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome, Cell 85 (6) (1996) 841–851. [DOI] [PubMed] [Google Scholar]
- [69].Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. , The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma, Nat. Genet 46 (5) (2014) 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. , Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma, Nature 482 (7384) (2012) 226–231. [DOI] [PubMed] [Google Scholar]
- [71].Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. , Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma, Cancer Cell 22 (4) (2012) 425–437. [DOI] [PubMed] [Google Scholar]
- [72].Lu N, Liu J, Xu M, Liang J, Wang Y, Wu Z, Xing Y, Diao F, CSMD3 is associated with tumor mutation burden and immune infiltration in ovarian cancer patients, Int. J. Gen. Med 14 (2021) 7647–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Noske A, Brandt S, Valtcheva N, Wagner U, Zhong Q, Bellini E, Fink D, Obermann EC, Moch H, Wild PJ, Detection of CCNE1/URI (19q12) amplification by in situ hybridisation is common in high grade and type II endometrial cancer, Oncotarget 8 (9) (2017) 14794–14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Noske A, Henricksen LA, LaFleur B, Zimmermann AK, Tubbs A, Singh S, Storz M, Fink D, Moch H, Characterization of the 19q12 amplification including CCNE1 and URI in different epithelial ovarian cancer subtypes, Exp. Mol. Pathol 98 (1) (2015) 47–54. [DOI] [PubMed] [Google Scholar]
- [75].Chiyoda T, Tsuda H, Tanaka H, Kataoka F, Nomura H, Nishimura S, Takano M, Susumu N, Saya H, Aoki D, Expression profiles of carcinosarcoma of the uterine corpus-are these similar to carcinoma or sarcoma? Genes Chromosomes Cancer 51 (3) (2012) 229–239. [DOI] [PubMed] [Google Scholar]
- [76].Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, et al. , Common variants at 19p13 are associated with susceptibility to ovarian cancer, Nat. Genet 42 (10) (2010) 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, et al. , Integrated molecular analysis of adult T cell leukemia/lymphoma, Nat. Genet 47 (11) (2015) 1304–1315. [DOI] [PubMed] [Google Scholar]
- [78].Matsuoka M, Jeang KT, Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation, Nat. Rev. Cancer 7 (4) (2007) 270–280. [DOI] [PubMed] [Google Scholar]
- [79].Yamagishi M, Watanabe T, Molecular hallmarks of adult T cell leukemia, Front. Microbiol 3 (2012) 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Nakagawa M, Schmitz R, Xiao W, Goldman CK, Xu W, Yang Y, Yu X, Waldmann TA, Staudt LM, Gain-of-function CCR4 mutations in adult T cell leukemia/lymphoma, J. Exp. Med 211 (13) (2014) 2497–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. , The genetic basis of early T-cell precursor acute lymphoblastic leukaemia, Nature 481 (7380) (2012) 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, et al. , Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia, Nat. Commun 6 (2015) 6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. , Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia, Nature 475 (7354) (2011) 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. , Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion, Nat. Genet 43 (10) (2011) 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. , Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators, Nat. Genet 44 (7) (2012) 760–764. [DOI] [PubMed] [Google Scholar]
- [86].Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome, Nat. Genet 39 (3) (2007) 311–318. [DOI] [PubMed] [Google Scholar]
- [87].Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. , Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer, Nat. Genet 44 (10) (2012) 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wu K, Zhang X, Li F, Xiao D, Hou Y, Zhu S, Liu D, Ye X, Ye M, Yang J, et al. , Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas, Nat. Commun 6 (2015) 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Scarlett CJ, Salisbury EL, Biankin AV, Kench J, Precursor lesions in pancreatic cancer: morphological and molecular pathology, Pathology 43 (3) (2011) 183–200. [DOI] [PubMed] [Google Scholar]
- [90].Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. , The genomic complexity of primary human prostate cancer, Nature 470 (7333) (2011) 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Decker B, Karyadi DM, Davis BW, Karlins E, Tillmans LS, Stanford JL, Thibodeau SN, Ostrander EA, Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors, Am. J. Hum. Genet 98 (5) (2016) 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. , Melanoma genome sequencing reveals frequent PREX2 mutations, Nature 485 (7399) (2012) 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. , A comprehensive catalogue of somatic mutations from a human cancer genome, Nature 463 (7278) (2010) 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. , Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors, Cancer Discov. 4 (2) (2014) 216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. , The mutational landscape of head and neck squamous cell carcinoma, Science 333 (6046) (2011) 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cazier JB, Rao SR, McLean CM, Walker AK, Wright BJ, Jaeger EE, Kartsonaki C, Marsden L, Yau C, Camps C, et al. , Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden, Nat. Commun 5 (2014) 3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE, Recent segmental duplications in the human genome, Science 297 (5583) (2002) 1003–1007. [DOI] [PubMed] [Google Scholar]
- [98].Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE, Segmental duplications: organization and impact within the current human genome project assembly, Genome Res. 11 (6) (2001) 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yan H, Bigner DD, Velculescu V, Parsons DW, Mutant metabolic enzymes are at the origin of gliomas, Cancer Res. 69 (24) (2009) 9157–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. , Cancer-associated IDH1 mutations produce 2-hydroxyglutarate, Nature 462 (7274) (2009) 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Pappula AL, Rasheed S, Mirzaei G, Petreaca RC, Bouley RA, A genome-wide profiling of glioma patients with an IDH1 mutation using the catalogue of somatic mutations in cancer database, Cancers (Basel) 13 (17) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Citron ML, Berry DA, Cirrincione C, Hudis C, Winer EP, Gradishar WJ, Davidson NE, Martino S, Livingston R, Ingle JN, et al. , Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741, J. Clin. Oncol 21 (8) (2003) 1431–1439. [DOI] [PubMed] [Google Scholar]
- [103].Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. , Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution, Genome Res. 17 (9) (2007) 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. , Complex landscapes of somatic rearrangement in human breast cancer genomes, Nature 462 (7276) (2009) 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. , Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line, Proc. Natl. Acad. Sci. U. S. A 106 (6) (2009) 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Davies V, Voutsadakis IA, Amplification of chromosome 17 centromere (CEP17) in breast cancer patients with a result of HER2 2+/- by immunohistochemistry, Cancer Invest. 38 (2) (2020) 94–101. [DOI] [PubMed] [Google Scholar]
- [107].Voutsadakis IA, Chromosome 17 centromere amplification and chromosomal instability (CIN) in breast cancer: pathogenic and therapeutic implications, Neoplasma 66 (6) (2019) 859–869. [DOI] [PubMed] [Google Scholar]
- [108].Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. , Genome-wide maps of chromatin state in pluripotent and lineage-committed cells, Nature 448 (7153) (2007) 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang Y, Chang JF, Sun J, Chen L, Yang XM, Tang HY, Jing YY, Kang X, He ZM, Wu JY, et al. , Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability, J. Cell. Sci 131 (12) (2018). [DOI] [PubMed] [Google Scholar]
- [110].Lhermitte B, Blandin AF, Coca A, Guerin E, Durand A, Entz-Werle N, Signaling pathway deregulation and molecular alterations across pediatric medulloblastomas, Neurochirurgie 67 (1) (2021) 39–45. [DOI] [PubMed] [Google Scholar]
- [111].Kwon E, Wang W, Tsai LH, Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets, Mol. Psychiatry 18 (1) (2013) 11–12. [DOI] [PubMed] [Google Scholar]
- [112].Shimizu A, Asakawa S, Sasaki T, Yamazaki S, Yamagata H, Kudoh J, Minoshima S, Kondo I, Shimizu N, A novel giant gene CSMD3 encoding a protein with CUB and sushi multiple domains: a candidate gene for benign adult familial myoclonic epilepsy on human chromosome 8q23.3-q24.1, Biochem. Biophys. Res. Commun 309 (1) (2003) 143–154. [DOI] [PubMed] [Google Scholar]
- [113].Ishitsuka K, Tamura K, Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma, Lancet Oncol. 15 (11) (2014) e517–526. [DOI] [PubMed] [Google Scholar]
- [114].Oka J, Matsumoto A, Hosokawa Y, Inoue S, Molecular cloning of human cytosolic purine 5’-nucleotidase, Biochem. Biophys. Res. Commun 205 (1) (1994) 917–922. [DOI] [PubMed] [Google Scholar]
- [115].Spychala J, Madrid-Marina V, Fox IH, High Km soluble 5’-nucleotidase from human placenta. Properties and allosteric regulation by IMP and ATP, J. Biol. Chem 263 (35) (1988) 18759–18765. [PubMed] [Google Scholar]
- [116].Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, Paietta E, Racevskis J, Rowe JM, Tallman MS, et al. , Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL, Nat. Med 19 (3) (2013) 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ramsay AJ, Martinez-Trillos A, Jares P, Rodriguez D, Kwarciak A, Quesada V, Next-generation sequencing reveals the secrets of the chronic lymphocytic leukemia genome, Clin. Transl. Oncol 15 (1) (2013) 3–8. [DOI] [PubMed] [Google Scholar]
- [118].Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, Rosolowski M, Ammerpohl O, Wagener R, Bernhart SH, et al. , Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing, Nat. Genet 44 (12) (2012) 1316–1320. [DOI] [PubMed] [Google Scholar]
- [119].Klapproth K, Wirth T, Advances in the understanding of MYC-induced lymphomagenesis, Br. J. Haematol 149 (4) (2010) 484–497. [DOI] [PubMed] [Google Scholar]
- [120].Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. , Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics, Nature 490 (7418) (2012) 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Chen HY, Bohlen JF, Maher BJ, Molecular and cellular function of transcription factor 4 in pitt-hopkins syndrome, Dev. Neurosci 43 (3–4) (2021) 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Waterman ML, Lymphoid enhancer factor/T cell factor expression in colorectal cancer, Cancer Metastasis Rev. 23 (1–2) (2004) 41–52. [DOI] [PubMed] [Google Scholar]
- [123].Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. , A small-cell lung cancer genome with complex signatures of tobacco exposure, Nature 463 (7278) (2010) 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Wistuba II, Gazdar AF, Minna JD, Molecular genetics of small cell lung carcinoma, Semin. Oncol 28 (2 Suppl. 4) (2001) 3–13. [PubMed] [Google Scholar]
- [125].Horowitz JM, Park SH, Bogenmann E, Cheng JC, Yandell DW, Kaye FJ, Minna JD, Dryja TP, Weinberg RA, Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells, Proc. Natl. Acad. Sci. U. S. A 87 (7) (1990) 2775–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Mori N, Yokota J, Akiyama T, Sameshima Y, Okamoto A, Mizoguchi H, Toyoshima K, Sugimura T, Terada M, Variable mutations of the RB gene in small-cell lung carcinoma, Oncogene 5 (11) (1990) 1713–1717. [PubMed] [Google Scholar]
- [127].Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, Sugio K, Smith DI, Liu W, PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers, Oncogene 17 (4) (1998) 475–479. [DOI] [PubMed] [Google Scholar]
- [128].Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR 3rd, Russell P, et al. , Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases, Cell 138 (1) (2009) 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW, Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair, Cell 138 (1) (2009) 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hsu E, Zemke NR, Berk AJ, Promoter-specific changes in initiation, elongation, and homeostasis of histone H3 acetylation during CBP/p300 inhibition, Elife (2021) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wilson A, Yakovlev VA, Cells redox environment modulates BRCA1 expression and DNA homologous recombination repair, Free Radic. Biol. Med 101 (2016) 190–201. [DOI] [PubMed] [Google Scholar]
- [132].Marini F, Giusti F, Tonelli F, Brandi ML, Pancreatic neuroendocrine neoplasms in multiple endocrine neoplasia type 1, Int. J. Mol. Sci 22 (8) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Nelakurti DD, Pappula AL, Rajasekaran S, Miles WO, Petreaca RC, Comprehensive analysis of MEN1 mutations and their role in cancer, Cancers (Basel) 12 (9) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. , Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate, Nat. Genet 41 (5) (2009) 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. , Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis, Nat. Genet 41 (5) (2009) 524–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. , Mutations of the BRAF gene in human cancer, Nature 417 (6892) (2002) 949–954. [DOI] [PubMed] [Google Scholar]
- [137].Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D, Springer CJ, Marais R, V599EB-RAF is an oncogene in melanocytes, Cancer Res. 64 (7) (2004) 2338–2342. [DOI] [PubMed] [Google Scholar]
- [138].Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, Hopkins B, Keniry M, Sulis ML, Mense S, et al. , Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a, Science 325 (5945) (2009) 1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kaseb H, Kuhn J, Babiker HM, Rhabdomyosarcoma. StatPearls, 2021. Treasure Island (FL). [Google Scholar]
- [140].Lan H, Sun Y, Tumor suppressor FBXW7 and its regulation of DNA damage response and repair, Front. Cell Dev. Biol 9 (2021), 751574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Huynh KD, Fischle W, Verdin E, Bardwell VJ, BCoR, a novel corepressor involved in BCL-6 repression, Genes Dev. 14 (14) (2000) 1810–1823. [PMC free article] [PubMed] [Google Scholar]
- [142].Linardic CM, PAX3-FOXO1 fusion gene in rhabdomyosarcoma, Cancer Lett. 270 (1) (2008) 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Barr FG, The role of chimeric paired box transcription factors in the pathogenesis of pediatric rhabdomysarcoma, Cancer Res. 59 (Suppl. 7) (1999) 1711s–1715s. [PubMed] [Google Scholar]
- [144].Leemans CR, Braakhuis BJ, Brakenhoff RH, The molecular biology of head and neck cancer, Nat. Rev. Cancer 11 (1) (2011) 9–22. [DOI] [PubMed] [Google Scholar]
- [145].Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S, Chromosome territories–a functional nuclear landscape, Curr. Opin. Cell Biol 18 (3) (2006) 307–316. [DOI] [PubMed] [Google Scholar]
- [146].Misteli T, Beyond the sequence: cellular organization of genome function, Cell 128 (4) (2007) 787–800. [DOI] [PubMed] [Google Scholar]
- [147].McCord RP, Balajee A, 3D genome organization influences the chromosome translocation pattern, Adv. Exp. Med. Biol 1044 (2018) 113–133. [DOI] [PubMed] [Google Scholar]
- [148].Gothe HJ, Minneker V, Roukos V, Dynamics of double-strand breaks: implications for the formation of chromosome translocations, Adv. Exp. Med. Biol 1044 (2018) 27–38. [DOI] [PubMed] [Google Scholar]
- [149].Anderson RM, Stevens DL, Goodhead DT, M-FISH analysis shows that complex chromosome aberrations induced by alpha -particle tracks are cumulative products of localized rearrangements, Proc. Natl. Acad. Sci. U. S. A 99 (19) (2002) 12167–12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R, Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains, Science 303 (5654) (2004) 92–95. [DOI] [PubMed] [Google Scholar]
- [151].Aymard F, Aguirrebengoa M, Guillou E, Javierre BM, Bugler B, Arnould C, Rocher V, Iacovoni JS, Biernacka A, Skrzypczak M, et al. , Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes, Nat. Struct. Mol. Biol 24 (4) (2017) 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lemaitre C, Grabarz A, Tsouroula K, Andronov L, Furst A, Pankotai T, Heyer V, Rogier M, Attwood KM, Kessler P, et al. , Nuclear position dictates DNA repair pathway choice, Genes Dev. 28 (22) (2014) 2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Richardson C, Moynahan ME, Jasin M, Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations, Genes Dev. 12 (24) (1998) 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T, Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages, J. Cell Biol 160 (5) (2003) 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Thomson I, Gilchrist S, Bickmore WA, Chubb JR, The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1, Curr. Biol 14 (2) (2004) 166–172. [DOI] [PubMed] [Google Scholar]
- [156].Haber JE, Leung WY, Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends, Proc. Natl. Acad. Sci. U. S. A 93 (24) (1996) 13949–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Tubbs A, Nussenzweig A, Endogenous DNA damage as a source of genomic instability in cancer, Cell 168 (4) (2017) 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M, Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics, Ann. Intern. Med 138 (10) (2003) 819–830. [DOI] [PubMed] [Google Scholar]
- [159].Lukasova E, Kozubek S, Kozubek M, Kjeronska J, Ryznar L, Horakova J, Krahulcova E, Horneck G, Localisation and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukaemia, Hum. Genet 100 (5–6) (1997) 525–535. [DOI] [PubMed] [Google Scholar]
- [160].Ghosh D, Raghavan SC, Nonhomologous end joining: new accessory factors fine tune the machinery, Trends Genet. 37 (6) (2021) 582–599. [DOI] [PubMed] [Google Scholar]
- [161].Elbakry A, Lobrich M, Homologous recombination subpathways: a tangle to resolve, Front. Genet 12 (2021), 723847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S, Differential usage of non-homologous end-joining and homologous recombination in double strand break repair, DNA Repair (Amst.) 5 (9–10) (2006) 1021–1029. [DOI] [PubMed] [Google Scholar]
- [163].Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. , Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining, Mol. Cell 55 (6) (2014) 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bhargava R, Onyango DO, Stark JM, Regulation of single-strand annealing and its role in genome maintenance, Trends Genet. 32 (9) (2016) 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang W, Edwards A, Fan W, Deininger P, Zhang K, Alu distribution and mutation types of cancer genes, BMC Genomics 12 (2011) 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Pavlicek A, Noskov VN, Kouprina N, Barrett JC, Jurka J, Larionov V, Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition, Hum. Mol. Genet 13 (22) (2004) 2737–2751. [DOI] [PubMed] [Google Scholar]
- [167].Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA, The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia, Proc. Natl. Acad. Sci. U. S. A 95 (5) (1998) 2390–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Sakofsky CJ, Malkova A, Break induced replication in eukaryotes: mechanisms, functions, and consequences, Crit. Rev. Biochem. Mol. Biol 52 (4) (2017) 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Lee JH, Paull TT, Cellular functions of the protein kinase ATM and their relevance to human disease, Nat. Rev. Mol. Cell Biol 22 (12) (2021) 796–814. [DOI] [PubMed] [Google Scholar]
- [170].Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. , ATM stabilizes DNA double-strand-break complexes during V(D)J recombination, Nature 442 (7101) (2006) 466–470. [DOI] [PubMed] [Google Scholar]
- [171].Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. , ATM prevents the persistence and propagation of chromosome breaks in lymphocytes, Cell 130 (1) (2007) 63–75. [DOI] [PubMed] [Google Scholar]
- [172].Yin B, Savic V, Bassing CH, ATM prevents unattended DNA double strand breaks on site and in generations to come, Cancer Biol. Ther 6 (12) (2007) 1837–1839. [DOI] [PubMed] [Google Scholar]
- [173].Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ, Kokkalis A, Roumelioti FM, Gagos S, Tzetis M, et al. , Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing, Nat. Cell Biol 18 (7) (2016) 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Galanos P, Pappas G, Polyzos A, Kotsinas A, Svolaki I, Giakoumakis NN, Glytsou C, Pateras IS, Swain U, Souliotis VL, et al. , Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability, Genome Biol. 19 (1) (2018) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Panagopoulos I, Heim S, Interstitial deletions generating fusion genes, Cancer Genomics Proteomics 18 (3) (2021) 167–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Knudson AG Jr., Mutation and cancer: statistical study of retinoblastoma, Proc. Natl. Acad. Sci. U. S. A 68 (4) (1971) 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Santarosa M, Ashworth A, Haploinsufficiency for tumour suppressor genes: when you don’t need to go all the way, Biochim. Biophys. Acta 1654 (2) (2004) 105–122. [DOI] [PubMed] [Google Scholar]
- [178].Quon KC, Berns A, Haplo-insufficiency? Let me count the ways, Genes Dev. 15 (22) (2001) 2917–2921. [DOI] [PubMed] [Google Scholar]
- [179].Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ, The murine gene p27Kip1 is haplo-insufficient for tumour suppression, Nature 396 (6707) (1998) 177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Komuro H, Valentine MB, Rubnitz JE, Saito M, Raimondi SC, Carroll AJ, Yi T, Sherr CJ, Look AT, p27KIP1 deletions in childhood acute lymphoblastic leukemia, Neoplasia 1 (3) (1999) 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Knudson AG, Two genetic hits (more or less) to cancer, Nat. Rev. Cancer 1 (2) (2001) 157–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.