Fig 1.
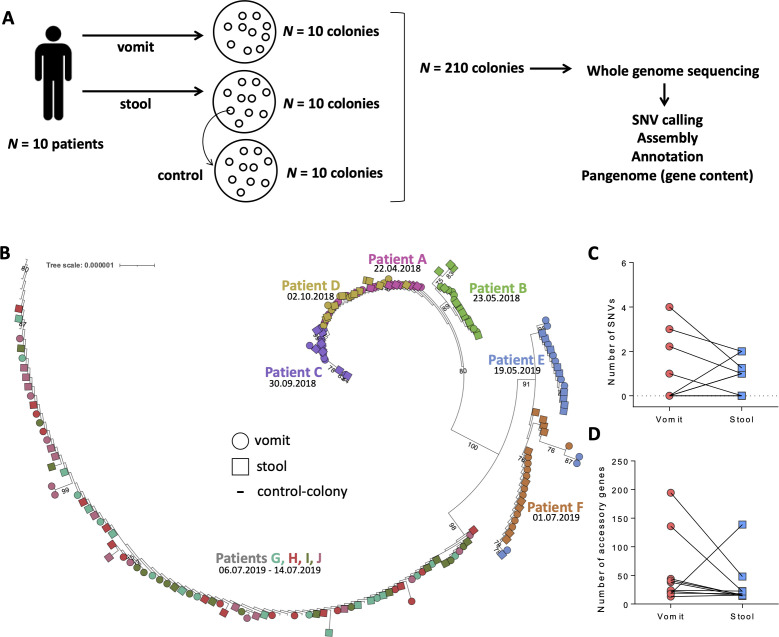
V. cholerae O1 within-patient diversity is modestly reduced in stool compared to vomit. (A) Ten patients were recruited for this study. For each patient, vomit and stool samples were plated to isolate colonies. From each vomit and stool sample, 10 colonies were used for whole-genome sequencing, in addition to 10 control colonies to control for sequencing errors. Sequence data were processed independently for single-nucleotide variant (SNV) calling and pangenome analysis (gene presence/absence) after assembly and annotations, with control colonies used to set filtering thresholds. (B) Maximum-likelihood phylogeny of 200 isolates sequenced, demonstrating that isolates cluster by patient and collection date. Ten control colony isolates are also shown. The tree is rooted on the MJ-1236 V. cholerae O1 reference genome (branching at the base of patient C). Samples are colored per patient. Collection dates are indicated below patient identifiers. Bootstrap values >75 are displayed. (C) Intra-patient variation based on SNVs called against the MJ-1236 reference genome across paired vomit and stool V. cholerae genomes, demonstrating a decrease in SNVs in four patients, an increase in three patients, and no change in three patients. The number of SNVs is normalized per 10 colonies, to account for some patients yielding only nine sequenced colonies. (D) Comparison of within-patient V. cholerae gene content in vomit and stool from 10 patients, showing a decrease in 8 patients and an increase in 2 patients. Only “accessory” genes that vary in their presence/absence in our data set are counted here; “core” genes common to all genomes are not included.