Abstract
In August 2014, a low-pathogenic H7N3 influenza A virus was isolated from pheasants at a New Jersey gamebird farm and hunting preserve. In this study, we use phylogenetic analyses and calculations of genetic similarity to gain inference into the genetic ancestry of this virus and to identify potential routes of transmission. Results of maximum-likelihood (ML) and maximum-clade-credibility (MCC) phylogenetic analyses provide evidence that A/pheasant/New Jersey/26996–2/2014 (H7N3) had closely related H7 hemagglutinin (HA) and N3 neuraminidase (NA) gene segments as compared to influenza A viruses circulating among wild waterfowl in the central and eastern USA. The estimated time of the most recent common ancestry (TMRCA) between the pheasant virus and those most closely related from wild waterfowl was early 2013 for both the H7 HA and N3 NA gene segments. None of the viruses from waterfowl identified as being most closely related to A/pheasant/New Jersey/26996–2/2014 at the HA and NA gene segments in ML and MCC phylogenetic analyses shared ≥99 % nucleotide sequence identity for internal gene segment sequences. This result indicates that specific viral strains identified in this study as being closely related to the HA and NA gene segments of A/pheasant/New Jersey/26996–2/2014 were not the direct predecessors of the etiological agent identified during the New Jersey outbreak. However, the recent common ancestry of the H7 and N3 gene segments of waterfowl-origin viruses and the virus isolated from pheasants suggests that viral diversity maintained in wild waterfowl likely played an important role in the emergence of A/pheasant/New Jersey/26996–2/2014.
Introduction
Wild aquatic birds, specifically waterfowl, gulls, and shorebirds, maintain the majority of the antigenic and genetic diversity of influenza A viruses (IAVs) [1]. Of the 16 hemagglutinin (HA) subtypes identified in wild birds, two, H5 and H7, have previously developed high pathogenicity in poultry through the accumulation of mutations at the HA cleavage site. Viruses of these subtypes are therefore of agricultural concern, and poultry infections caused by IAVs of the H5 and H7 subtypes, regardless of pathogenicity, are reportable to the World Organization for Animal Health (OIE). As such, introductions of H5 and H7 IAVs into poultry production systems may lead to economic losses, either directly through morbidity and mortality, or indirectly through the application of control and eradication measures or via restrictions in trade. Additionally, spillover of poultry-origin IAVs of the H5 and H7 subtypes has resulted in human infections, raising concerns regarding the pandemic potential of such viruses [2–4]. Therefore, information leading to a better understanding of viral transmission across the wild bird-poultry interface may be useful for improving biosecurity practices that minimize the frequency of introductions of H5 and H7 subtype IAVs into domestic birds, thereby promoting population health.
Although H5 and H7 IAVs have been detected in commercial poultry, live-bird markets, and domestically reared game birds in the USA numerous times in recent decades [5–8], only a limited number of studies have identified molecular or epidemiological evidence for transmission of H5 and H7 subtype IAVs across the wild bird-poultry interface. Low-pathogenic H7N9 IAVs detected in Minnesota, USA turkey farms during 2009–2011 were closely related to viruses circulating among wild waterfowl in the Mississippi Flyway [9]. Similarly, highly pathogenic H5N2 viruses detected in poultry in British Columbia, Canada, in 2014, and subsequently the Midwestern states of the USA in 2015, also appeared to share common ancestry with IAVs detected in North American waterfowl [10–12]. Molecular and epidemiological links between other H5 and H7 IAVs detected in domestic birds in the USA and the wild bird reservoir are generally less clear, but evidence suggests that spillover of IAVs from the wild bird reservoir into domestic poultry may not be uncommon [13].
In late August 2014, routine testing of pheasants (family Phasianidae, subfamily Phasianinae) for IAVs was conducted on a New Jersey, USA gamebird farm and hunting preserve. There had been no clinical signs or increased mortality reported among the holdings of approximately 7200 pheasants and 44,000 mallards (Anas platyrhynchos). Three pooled samples of 10 oropharyngeal swabs collected from pheasants were tested by the United States Department of Agriculture Animal and Plant Health Inspection Service National Veterinary Services Laboratory (NVSL), resulting in the isolation of an H7N3 subtype IAV predicted to be low pathogenic in poultry based on deduced amino acid motifs at the fusion cleavage site. Control measures, including quarantine, were applied to the gamebird farm and hunting preserve, and follow-up testing of the affected premises and an epidemiologically linked farm suggested that the outbreak was contained [14]. However, the origins of the H7N3 IAV detected at the gamebird farm and hunting preserve were not determined.
In this investigation, we used genetic evidence to assess whether North American waterfowl, gulls, shorebirds, or poultry were the likely source of the H7N3 IAV that was introduced to New Jersey pheasants. Specifically, we used phylogenetic methods to assess the genetic ancestry of the H7 and N3 IAV gene segments and to estimate when the introduction of IAV into New Jersey pheasants may have occurred. Furthermore, we used genetic information to further assess the evidence for hitchhiking of internal gene segments with surface glycoproteins in the formation of the genomic constellation that was ultimately detected in New Jersey pheasants. Resulting information from this investigation may be useful for identifying potential routes of transmission across the wild bird-poultry interface, which may be used to improve biosecurity on game farms and hunting preserves in the USA and abroad.
Materials and methods
Influenza virus strain A/pheasant/New Jersey/26996–2/2014 (H7N3) was isolated by NVSL using standard protocols for RNA extraction, virus detection and quantitation by real-time RT-PCR, influenza virus subtype identification by molecular methods, and virus isolation, propagation, and titration in embryonated chicken eggs [15]. Viral RNA was extracted from the resultant isolate using a MagMAX Viral RNA Isolation Kit (Ambion/ThermoFisher Scientific). Complementary DNA was synthesized by reverse transcription using SuperScript III (Invitrogen/ThermoFisher Scientific). All eight gene segments of isolates were amplified by PCR, and complete genome sequencing was conducted using the Ion Torrent (Life Technologies) platform. Briefly, the PCR product was purified and a DNA library was prepared for the Ion Torrent using an IonXpress Plus Fragment Library Kit (Life Technologies) with Ion Xpress barcode adapters. The prepared library was quantitated using a Bioanalyzer DNA 1000 Kit. The quantitated library was diluted and pooled for amplification using the Ion One Touch 2 and ES systems. Following enrichment, DNA was loaded onto an Ion 314 or Ion 316 chip and sequenced using an Ion PGM 200 v2 Sequencing Kit. De novo and directed assembly of the genome sequence was carried out using the SeqMan NGen v4 program. Nucleotide sequences for the complete genome of A/pheasant/New Jersey/26996–2/2014 (H7N3) were deposited in the GenBank database under accession numbers KU740201–KU740208.
To investigate the genetic origins of A/pheasant/New Jersey/26996–2/2014 (H7N3), we obtained sequences for the complete coding region for all H7 HA and N3 neuraminidase (NA) gene segments previously reported from North America during 2000–2014 as available in the GenBank public database [16] (accessed 22–24 September 2015; n = 487 and n = 394, respectively). We also obtained genetic information for H7 HA (n = 14) and N3 NA (n = 13) gene segments for 20 additional viruses isolated from paired oropharyngeal/cloacal swab samples collected from live-captured blue-winged teal (Anas discors) in Texas (n = 1) and Louisiana (n = 15) during March 2014 and ruddy turnstones (Arenaria interpres) at Delaware Bay, New Jersey (n = 4) in May of 2014 (GenBank accession numbers: KT887256–KT887422; Supplemental Table 1). Viruses were isolated according to methods reported by Stallknecht et al. [17], and their genes were sequenced following protocols reported by Ramey et al. [18].
Maximum-likelihood (ML) phylogenies were first reconstructed in MEGA version 6.06 [19] to assess the genetic relationships among H7 HA and N3 NA gene segments for A/pheasant/New Jersey/26996–2/2014 (H7N3), isolates derived from blue-winged teal and ruddy turnstones, and IAVs previously reported from North America during 2000–2014 using the Nucleotide: Nearest-Neighbor-Inter-change method, with 1000 bootstrap replicates (n = 501 and 407 for H7 HA and N3NA gene segment sequences, respectively). Subsequently, genetic ancestry for strongly supported clades (bootstrap support values ≥70) containing sequence information for the H7 HA (n = 65 sequences in clade) and N3 NA (n = 112 sequences in clade) gene segments A/pheasant/New Jersey/26996–2/2014 (H7N3) was further investigated by reconstructing maximum-clade-credibility (MCC) phylogenetic trees.
Bayesian Markov Chain Monte Carlo coalescent analysis was performed using BEAST 1.7.4 [20]. The uncorrelated exponential molecular clock was selected after comparison of Bayes factors with estimates obtained with the strict clock and uncorrelated log-normal clocks. The SRD06 nucleotide substitution model [21] and a Bayesian skyline coalescent tree prior were used in all simulations [22]. We performed two independent analyses with chain lengths of 80 million generations sampled every 1000 iterations, and the first 20 % of the trees were discarded as burn-in.
To obtain information about the hitchhiking of internal gene segments with surface glycoproteins, we calculated nucleotide pairwise distances (PWD) for complete coding region genetic sequences of internal gene segments between A/pheasant/New Jersey/26996–2/2014 (H7N3) and other IAV strains sharing ≥99 % nucleotide sequence identity at either the HA or NA gene segment. Shared nucleotide sequence identity of ≥99 % between A/pheasant/New Jersey/26996–2/2014 (H7N3) and other IAV strains in internal gene segments was taken as evidence for hitchhiking of internal genes with surface glycoproteins in the formation of the genomic constellation detected in New Jersey pheasants.
Results
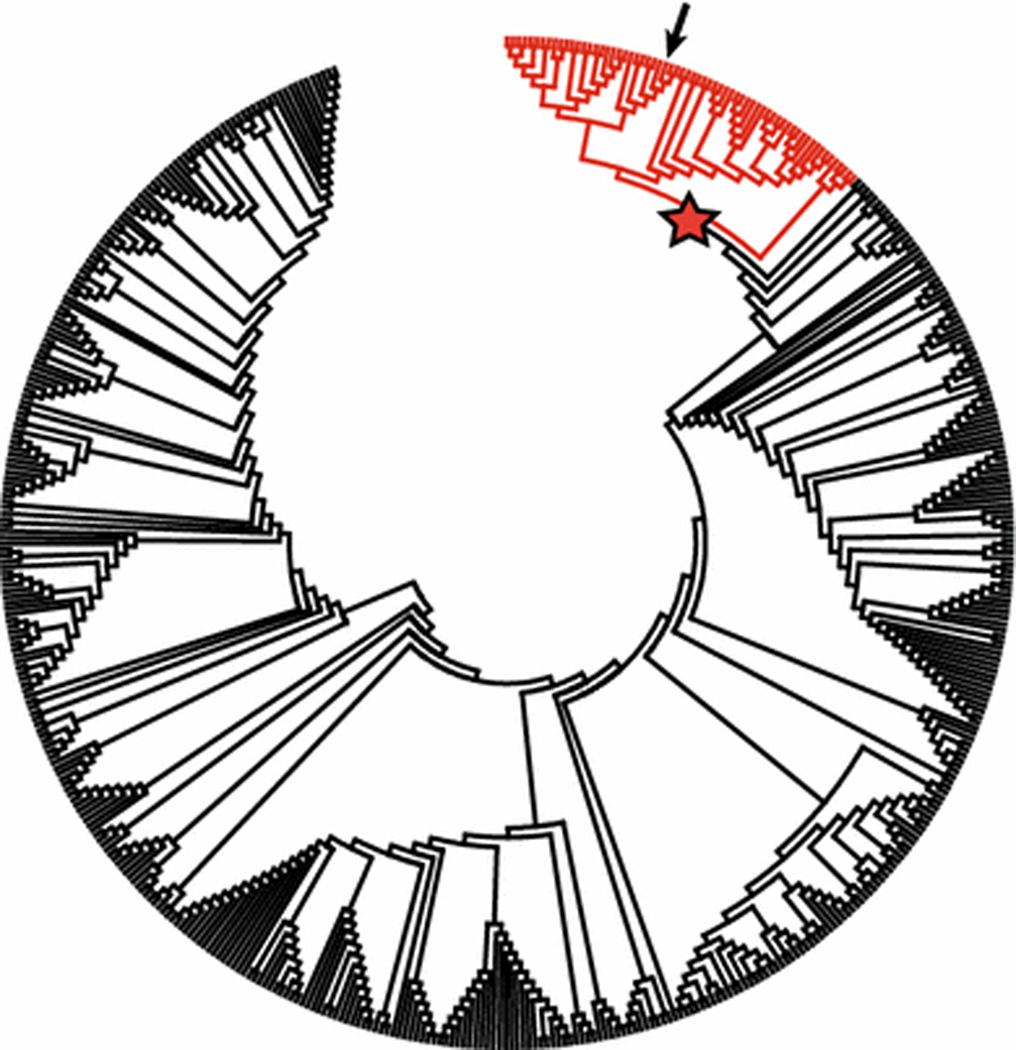
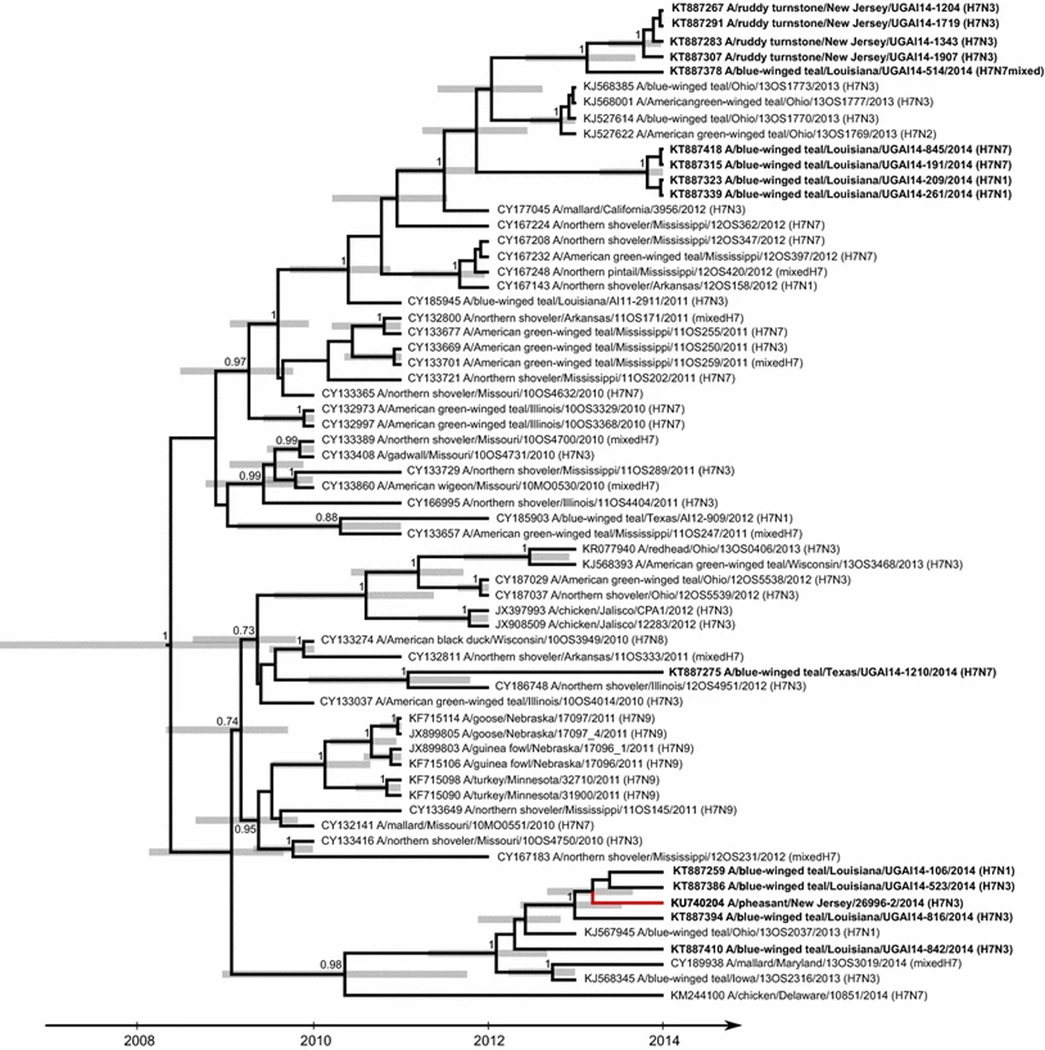
Using ML phylogenetic analysis, the genetic sequence for the H7 HA gene for A/pheasant/New Jersey/26996–2/2014 (H7N3) was nested within a strongly supported clade (bootstrap support value of 95) of 65 sequences derived from IAVs isolated from wild waterfowl, shorebirds, and domestic birds sampled in the USA and Mexico, including all 14 H7 subtype IAVs from blue-winged teal and ruddy turnstones sequenced in this study (Fig. 1, Supplemental Figure S1). The coalescent analysis performed for this clade provided more-detailed information on the possible origins of A/pheasant/New Jersey/26996–2/2014 (H7N3). The phylogenetic structure of the resulting tree (Fig. 2) revealed that the H7 HA gene segment of the virus isolated from a pheasant was closely related to those of viruses isolated from blue-winged teal (H7N1 and H7N3), sampled in Louisiana during 2014, with a time of the most recent common ancestry (TMRCA) estimated to be early 2013 (TMRCA; ±95 % highest posterior density [HPD]: 2013.2; 2012.7–2013.7). These viruses clustered together with other H7 viruses isolated from waterfowl sampled in Louisiana, Ohio, Maryland, and Iowa sampled in 2013–2014, with TMRCA estimated to be 2012 (2012; 2011.3–2012.7). These viruses formed an independent lineage in the phylogenetic tree that also included an H7N7 virus isolated from a chicken in Delaware in 2014, but with which the estimated TMRCA was earlier (2010.3; 2008.9–2011.8). Four H7 HA sequences, all obtained from IAVs isolated from blue-winged teal, shared ≥99 % nucleotide sequence identity with the HA sequence of A/pheasant/New Jersey/26996–2/2014 (H7N3) in PWD comparisons (Table 1). However, PWD comparisons for NA and internal gene segment sequences between A/pheasant/New Jersey/26996–2/2014 (H7N3) and four IAV isolates from blue-winged teal with highly similar HA gene sequences revealed lower levels of nucleotide sequence similarity (≤98 %; Table 1).
Fig. 1.
Maximum-likelihood phylogenetic tree depicting the inferred topology for 501 sequences of H7 hemagglutinin gene segments derived from influenza A viruses isolated from wild and domestic birds in North America. Branch tips for sequences selected for maximum-clade-credibility analysis are colored red. The bootstrap support value of the node indicated by a star is 95. The position of the sequence for A/pheasant/New Jersey/26996–2/2014 (H7N3) is indicated by an arrow. The expanded tree with complete tip labels (GenBank accession number and strain name) and bootstrap support values is available as supplemental material (Supplemental Figure S1)
Fig. 2.
Maximum-clade-credibility tree depicting the inferred ancestry of 65 full-length sequences for H7 hemagglutinin gene segments derived from influenza A viruses isolated from wild and domestic birds in North America. Posterior probability values ≥0.7 are reported. Grey bars indicate the 95 % HPD for the time of the most recent common ancestors. Sequences determined this study are indicated in bold. The branch tip for A/pheasant/New Jersey/26996–2/014 (H7N3) is colored red
Table 1.
Nucleotide sequence similarity by gene segment between A/pheasant/New Jersey/26996-2/2014 (H7N3) and other viruses with highly similar (≥99 %) genetic sequences for surface glycoproteins. Shared nucleotide sequence identity values ≥99 % are indicated in italics. Isolates sequenced as part of this study are indicated in bold font
| Strain | Surface glycoproteins | Internal gene segments | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| HA | NA | PB2 | PB1 | PA | NP | M | NS | |
| A/blue-winged teal/Iowa/13OS2316/2013 (H7N3) | 99 % | 98 % | 91 % | 96 % | 90 % | 94 % | 97 % | NC† |
| A/blue-winged teal/Louisiana/UGAI14-106/2014 (H7N1) | 99 % | NC† | 95 % | 95 % | 90 % | 94 % | 97 % | 96 % |
| A/blue-winged teal/Louisiana/UGAI14-478/2014 (H10N3) | NC† | 99 % | 91 % | 96 % | 90 % | 98 % | 98 % | 96 % |
| A/blue-winged teal/Louisiana/UGAI14-507/2014 (H10N3) | NC† | 99 % | 91 % | 96 % | 90 % | 98 % | 98 % | 93 % |
| A/blue-winged teal/Louisiana/UGAI14-523/2014 (H7N3) | 99 % | 94 % | 96 % | 95 % | 88 % | 94 % | 98 % | 97 % |
| A/blue-winged teal/Louisiana/UGAI14-816/2014 (H7N3) | 99 % | 95 % | 96 % | 94 % | 88 % | 93 % | 98 % | 96 % |
| A/blue-winged teal/Louisiana/UGAI14-842/2014 (H7N3) | 98 % | 99 % | 91 % | 96 % | 88 % | 94 % | 98 % | 96 % |
NC no comparison on account of different subtype or allele as compared to A/pheasant/New Jersey/26996-2/2014 (H7N3)
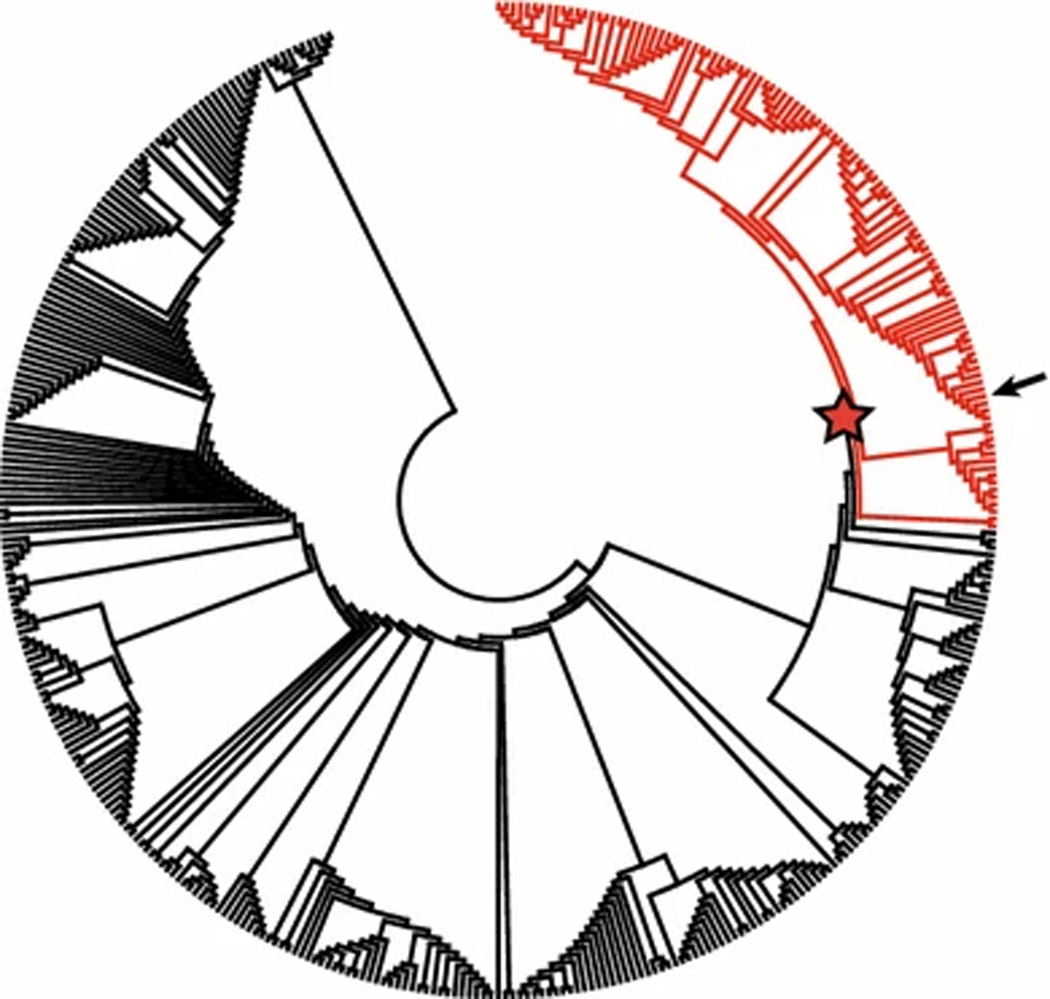
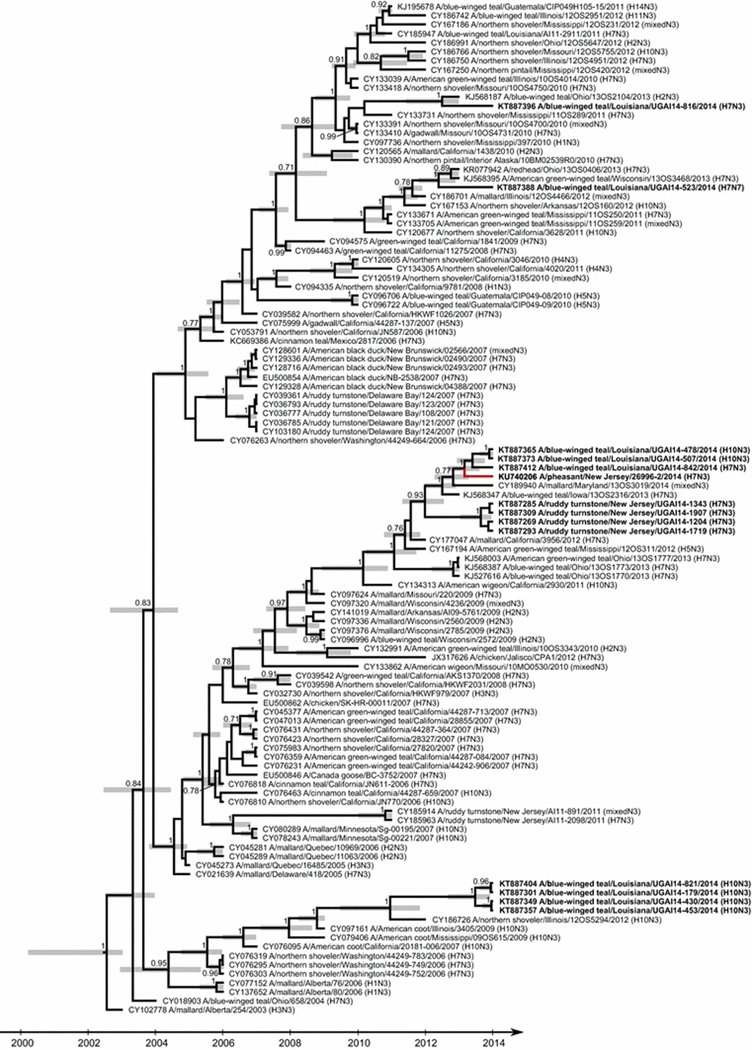
ML phylogenetic analysis supported inclusion of the N3 NA gene sequence of A/pheasant/New Jersey/26996–2/2014 (H7N3) within a strongly supported clade (bootstrap support value of 77) of 112 sequences derived from IAVs isolated from waterfowl, shorebird, and chicken samples collected in the USA, Canada, Mexico, and Guatemala, including 13 N3 subtype IAVs from blue-winged teal and ruddy turnstones sequenced for this study (Fig. 3, Supplemental Figure S2). The coalescent analysis performed for the N3 NA sequences revealed that the N3 NA gene segment of A/pheasant/New Jersey/26996–2/2014 was closely related to viruses isolated from blue-winged teal sampled in Louisiana during 2014 (Fig. 4) with an estimated TMRCA of early 2013 (2013.1; 2012.7–2013.6). These N3 NA gene segment sequences clustered together with those of other N3 IAVs isolated from waterfowl sampled in Maryland and Iowa in 2013 and 2014 (2012; 2011.3–2012.5). The majority of the N3 NA genetic sequences included in MCC analysis (110 of 112) were clustered into a single clade estimated to share common ancestry in 2004 (2004.4; 2003.8–2004.9). The N3 NA sequences for three isolates from blue-winged teal sequenced as part of this study shared ≥99 % nucleotide sequence identity with the NA sequence of A/pheasant/New Jersey/26996–2/2014 (H7N3); however, PWD comparisons between the isolates from blue-winged teal and the IAV from a New Jersey pheasant revealed shared nucleotide sequence identity ≤98 % for the HA gene and internal gene segments (Table 1).
Fig. 3.
Maximum-likelihood phylogenetic tree depicting the inferred topology for 407 sequences of N3 neuraminidase gene segments derived from influenza A viruses isolated from wild and domestic birds in North America. Branch tips for sequences selected for maximum-clade-credibility analysis are colored red. The bootstrap support value of the node indicated by a star is 77. The position of the sequence for A/pheasant/New Jersey/26996–2/2014 (H7N3) is indicated by an arrow. The expanded tree with complete tip labels (GenBank accession number and strain name) and bootstrap support values is available as supplemental material (Supplemental Figure S2)
Fig. 4.
Maximum-clade-credibility tree depicting the inferred ancestry of 112 full-length sequences for N3 neuraminidase gene segments derived from influenza A viruses isolated from wild and domestic birds in North America. Posterior probability values >0.7 are reported. Grey bars indicate the 95 % HPD for the time of the most recent common ancestors. Sequences determined in this study are indicated in bold. The branch tip for A/pheasant/New Jersey/26996–2/2014 (H7N3) is colored red
Discussion
Previous investigations have provided evidence for shared ancestry of IAVs detected in domestically reared birds in the USA with those isolated from samples collected from wild North American waterfowl [9–12]. Therefore, it is unsurprising that we found evidence for relatively recent common ancestry of H7 HA and N3 NA gene segments of A/pheasant/New Jersey/26996–2/2014 with viruses circulating in dabbling ducks sampled throughout the central and eastern USA, particularly as some minor poultry species, such as pheasants, may be readily infected by wild-bird-origin IAVs [23], and the game farm on which the pheasant virus was detected had considerable holdings of mallards. However, none of the viruses identified in wild waterfowl as being closely related to A/pheasant/New Jersey/26996–2/2014 in ML and MCC phylogenetic analyses also shared high (≥99 %) nucleotide sequence similarity with any internal gene segment sequences of the pheasant isolate. Furthermore, post-hoc MCC analyses provided evidence that IAVs most closely related to A/pheasant/New Jersey/26996–2/2014 at the HA and NA gene segments were not the most closely related viruses to the H7N3 pheasant virus at internal gene segments, nor was there support for recent common ancestry for internal gene segments of A/pheasant/New Jersey/26996–2/2014 and IAV strains sequenced for this study (Supplemental Figures S3–S8). Thus, although we have identified wild waterfowl as a putative source for the virus detected in New Jersey pheasants, we have not identified the direct predecessor virus strain.
Given that the TMRCA for both the H7 HA and N3 NA gene segments between A/pheasant/New Jersey/26996–2/2014 and the most closely related IAVs from waterfowl was estimated to be more than a year prior to viral detection in pheasants and that we were not able to identify a direct predecessor virus, rigorous fine-scale inference regarding the timing of viral introduction in the New Jersey bird-rearing facility is not currently possible. Furthermore, a lack of information on the IAV infection status of mallards at the New Jersey facility from which this H7N3 virus was isolated precludes inference regarding the role of captive-reared waterfowl in the emergence of A/pheasant/New Jersey/26996–2/2014. Additional sequence information from wild and/or domestic birds sampled in the USA during 2013–2014 may, however, allow more precise estimates of the time of introduction.
Our results build on the growing body of literature supporting viral diversity maintained in wild waterfowl as playing a particularly important role in the emergence of IAVs in poultry as compared to shorebirds and gulls, other avian taxa typically identified as comprising the wild bird reservoir of IAVs [24]. Thus, biosecurity for domestically reared birds may be most efficiently optimized by focusing on interfaces between wild waterfowl (and associated wetland habitats) and poultry. For example, raising pheasants in close proximity to pen-reared waterfowl may promote viral exchange among taxa, whereas separate facilities with protocols in place to minimize viral spread via fomites and watering systems may greatly reduce the likelihood of interspecies transmission. Even though the detection of A/pheasant/New Jersey/26996–2/2014 was in a modest-sized game bird farm and the virus was predicted to be of low pathogenicity based on genetic sequencing, this event exemplifies a potential route through which IAVs may spill over from wild birds into domestic production, posing an elevated risk of disease to nearby commercial operations or those linked via trade.
Finally, in both the HA and NA trees, isolates derived from blue-winged teal and sequenced for this study displayed considerable genetic diversity with only a portion inferred as sharing recent common ancestry with A/pheasant/New Jersey/26996–2/2014. All of these blue-winged teal isolates originated from Texas and Louisiana as part of a single spring collection effort. This observed genetic diversity highlights the value of sequencing multiple isolates of the same subtype recovered from wild bird sample collections in order to reliably assess the ancestry and time of introduction of IAVs infecting domestic poultry.
Supplementary Material
Acknowledgements
We appreciate field and laboratory support by Nick Davis-Fields, Alinde Fojtik, Clara Kienzle, and John Reed. We thank John Pearce, Mary Pantin-Jackwood, and two anonymous reviewers for providing critical reviews.
Compliance with ethical standards
This project was Funded by the U.S. Geological Survey through the Wildlife Program of the Ecosystems Mission area, the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN272201400006C., and by ‘chaire mixte: institut national de la santé et de la recherche médicale – université de La Réunion’. None of the authors have any financial interests or conflict of interest with this article. Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00705-016-2947-z) contains supplementary material, which is available to authorized users.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claas ECJ, Osterhaus ADME, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG (1998) Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477 [DOI] [PubMed] [Google Scholar]
- 3.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Oseterhaus A, Bosman A (2004) Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587–593 [DOI] [PubMed] [Google Scholar]
- 4.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y (2013) Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897 [DOI] [PubMed] [Google Scholar]
- 5.Saito T, Horimoto T, Kawaoka Y, Senne DA, Webster RG (1994) Emergence of a potentially pathogenic H5N2 influenza virus in chickens. Virology 201:277–284 [DOI] [PubMed] [Google Scholar]
- 6.Suarez DL, Senne DA (2000) Sequence analysis of related low-pathogenic and highly pathogenic H5N2 avian influenza isolates from United States live bird markets and poultry farms from 1983 to 1989. Avian Dis 44:356–364 [PubMed] [Google Scholar]
- 7.Spackman E, Senne DA, Davison S, Suarez DL (2003) Sequence analysis of recent H7 avian influenza viruses associated with three different outbreaks in commercial poultry in the United States. J Virol 77:13399–13402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CW, Swayne DE, Linares JA, Senne DA, Suarez DL (2005) H5N2 avian influenza outbreak in Texas in 2004: the first highly pathogenic strain in the United States in 20 years? J Virol 79:11412–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebarbenchon C, Pedersen JC, Sreevatsan S, Ramey AM, Dugan VG, Halpin RA, Ferro PJ, Lupiani B, Enomoto S, Poulson RL, Smeltzer M, Cardona CJ, Tompkins SM, Wentwoth DE, Stallknecht DE, Brown JD (2015) H7N9 Influenza A virus in Turkeys in Minnesota. J Gen Virol 96:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Boschler V, Killian ML, Pedersen JC, Hines N, Gidlewski T, DeLiberto T, Sleeman JM (2015) Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis 21:886–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasick J, Berhane Y, Joseph T, Bowes V, Hisanaga T, Handel K, Alexandersen S (2014) Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada. Sci Rep 5:9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramey AM, Reeves AB, TeSlaa JL, Nashold S, Donnelly T, Bahl J, Hall JS (2016) Evidence for common ancestry among viruses isolated from wild birds in Beringia and highly pathogenic intercontinental reassortant H5N1 and H5N2 influenza A viruses. Infect Genet Evol 40:176–185 [DOI] [PubMed] [Google Scholar]
- 13.Lebarbenchon C, Stallknecht DE (2011) Host shifts and molecular evolution of H7 avian influenza virus hemagglutinin. Virol J 8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.OIE (2014) Follow-up report No. 3 on occurrence of low pathogenic avian influenza (poultry), United States of America. http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=16135. Accessed Nov 2015
- 15.Spackman E (ed) (2014) Animal influenza virus. Springer, New York [Google Scholar]
- 16.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D (2008) The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stallknecht DE, Shane SM, Zwank PJ, Senne DA, Kearney MT (1990) Avian influenza viruses from migratory and resident ducks of coastal Louisiana. Avian Dis 34:398–405 [PubMed] [Google Scholar]
- 18.Ramey AM, Reeves AB, Sonsthagen SA, TeSlaa JL, Nashold S, Donnelly T, Casler B, Hall JS (2015) Dispersal of H9N2 influenza A viruses between East Asia and North America by wild birds. Virology 482:79–83 [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapiro B, Rambaut A, Drummon AJ (2006) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23:7–9 [DOI] [PubMed] [Google Scholar]
- 22.Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22:1185–1192 [DOI] [PubMed] [Google Scholar]
- 23.Humberd J, Guan Y, Webster RG (2006) Comparison of the replication of influenza A viruses in Chinese ring-necked pheasants and chukar partridges. J Virol 80:2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krauss S, Stucker KM, Schobel SA, Danner A, Friedman K, Knowles JP, Kayali G, Niles LJ, Dey AD, Raven G, Pryor P (2015) Long-term surveillance of H7 influenza viruses in American wild aquatic birds: Are the H7N3 influenza viruses in wild birds the precursors of highly pathogenic strains in domestic poultry? Emerg Microbes Infect 4:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.