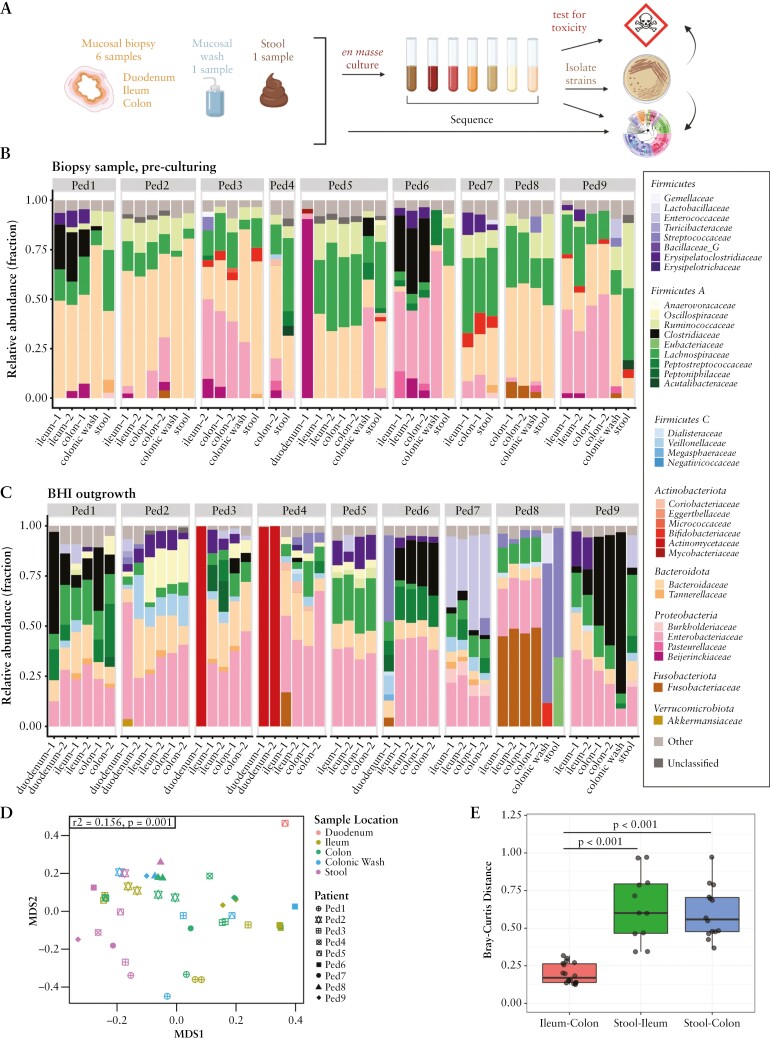
Figure 1.
Sample collection, workflow, and sequencing analysis from paediatric gastroenterology patients. [A] Sample workflow diagram. Patient samples sequenced [B] directly as biopsies pre-culturing and [C] after the second passage outgrowth in BHI media. Samples were sequenced using 16S V4 sequencing and classified at the family taxonomic rank. Minimum 10 000 reads filtering cutoff per sample. [D] Beta diversity analysis of the microbiome from outgrowth communities for each patient sample: NMDS ordination plot of Bray–Curtis distances. Colour represents sample location, shape represents patient. PERMANOVA was performed to test the impact of sample location on Bray–Curtis distance, stratified by patient [r2 = 0.156, p = 0.001]. [E] Boxplot of pairwise Bray–Curtis distances between intrapatient ileum, colon, and stool outgrowth communities. The p-values were determined by Wilcoxon rank sum test.