Figure 2.
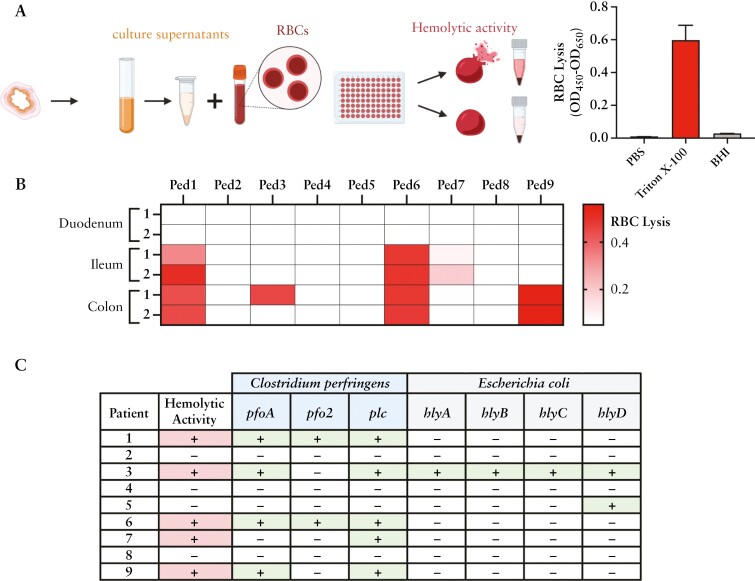
Patient biopsy community samples are haemolytic. [A] Experimental schematic for RBC lysis, measured as [OD450 − OD650]. Media control [BHI] and 100% lysis control [0.5% Triton X-100] are shown. [B] Biopsy outgrowth in BHI after the second passage for each patient was measured for RBC lysis. Two biopsies from each location were evaluated. [C] Long-read metagenome-assembled genomes [MAGs] of BHI outgrowth communities were searched against the virulence factor database [VFDB]. Presence of sequences homologous to known haemolysin genes is indicated with ‘+’ and absence with ‘-’. Haemolytic activity from samples outgrown in BHI is noted with ‘+’ and no activity with ‘-’. The genes pfoA, pfo2, plc, and hlyA are predicted haemolysins, and hlyB–D haemolysin accessory genes. Primary haemolytic data available in Supplementary Table 5.