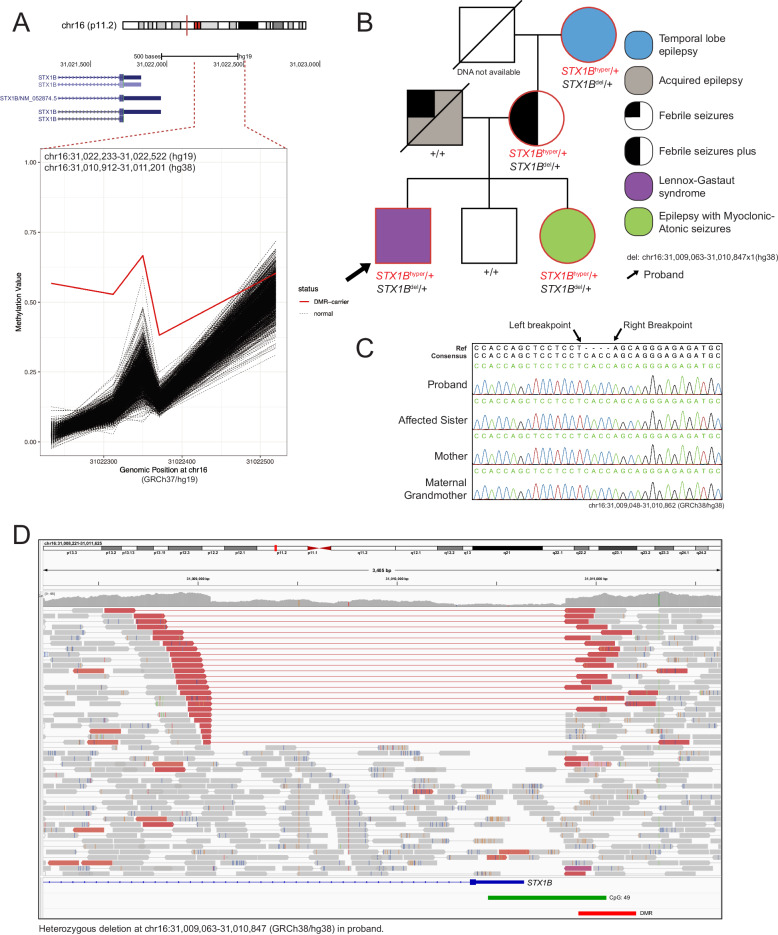
Fig. 4. Rare outlier DMR analysis identifies copy number deletion in a family with GEFs + .
A DMR plot depicting outlier hypermethylation of the STX1B promoter and TSS in a proband with uDEE detected through epivariation analysis. B Pedigree for the immediate family members indicating that the inheritance of the hypermethylation (red) and copy number deletion (detected through genome sequencing of the proband shown in Fig. 3D) resides on the maternal side. The arrow points to the proband. C Sanger sequencing validation of the copy number deletion breakpoints in the proband, affected sister, mother, and maternal grandmother. Inserted “CACC” sequence is present between the mapped breakpoints. The father and unaffected brother had no PCR product at the same reaction conditions, indicating they did not harbor the deletion. Primer pairs with one partner located within the deletion on each side of the breakpoint were used to amplify the wild-type allele as a control (Supplementary Fig. 13). D IGV view of genome sequencing data showing a 1784 bp heterozygous deletion encompassing part of intron 1, exon 1, the TSS, and the promoter of STX1B. The reads are colored by insertion size and pair orientation and viewed as pairs. The red pairs, which span the breakpoints of the deletion, indicate that the insertion size is greater than expected. The “CACC” insertion sequence is present in the soft clipped bases (not shown).