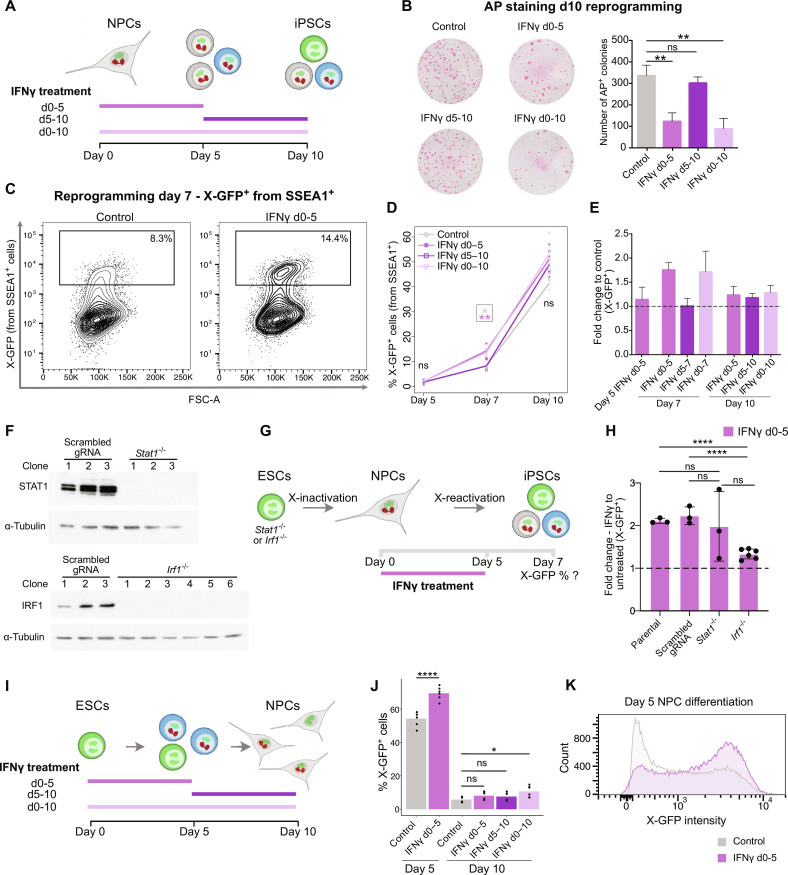
Fig. 2. IFNγ modulates colony formation and X-reactivation during reprogramming.
(A) For (B) to (E), IFNγ was added from day 0 to 5, day 5 to 10, day 0 to 10 (n = 3 reprogramming rounds). (B) AP stainings on day 10 of reprogramming (n = 3 for control, n = 4 for IFNγ treatments). Statistics (unpaired t tests): ns = nonsignificant; **P < 0.01. Error bars represent SD. (C) Flow cytometry plots of X-GFP+ (from SSEA1+) cells (gate) in control and IFNγ treatment (day 0 to 5) at day 7. (D) Flow cytometry analysis of X-GFP+ cells on reprogramming days 5, 7, and 10. Statistics (paired t tests): *P < 0.05; **P < 0.01. (E) Fold change of X-GFP percentages compared to each control. Error bars represent SD [calculations based on percentages in (D)]. (F) Western blot of STAT1 and IRF1 for three scrambled gRNA and three Stat1−/− clones (top) or six Irf1−/− clones (bottom). Loading control: ɑ-tubulin. (G) For (H), NPCs derived from three Stat1−/−, six Irf1−/−, three parental, and three scrambled gRNA ESC clones were reprogrammed into iPSCs ± IFNγ (day 0 to 5). X-GFP percentages measured by flow cytometry at day 7. (H) Fold change of X-GFP+ percentage (IFNγ versus untreated controls) on day 7 measured by flow cytometry. Clones with the same genotype listed in (G) are grouped; dots represent the mean of three technical replicates for each clone. Statistics (unpaired t tests): ****P < 0.0001. Error bars represent SD. (I) For (J) and (K), IFNγ treatment was done during NPC differentiation (day 0 to 5, day 5 to 10, and day 0 to 10) (n = 6 independent replicates). (J) X-GFP percentage on days 5 and 10 of NPC differentiation measured by flow cytometry. Statistics (paired t tests): *P < 0.05; ****P < 0.0001. (K) Flow cytometry histogram of X-GFP intensity in representative samples of control and IFNγ-treated day 5 NPCs.