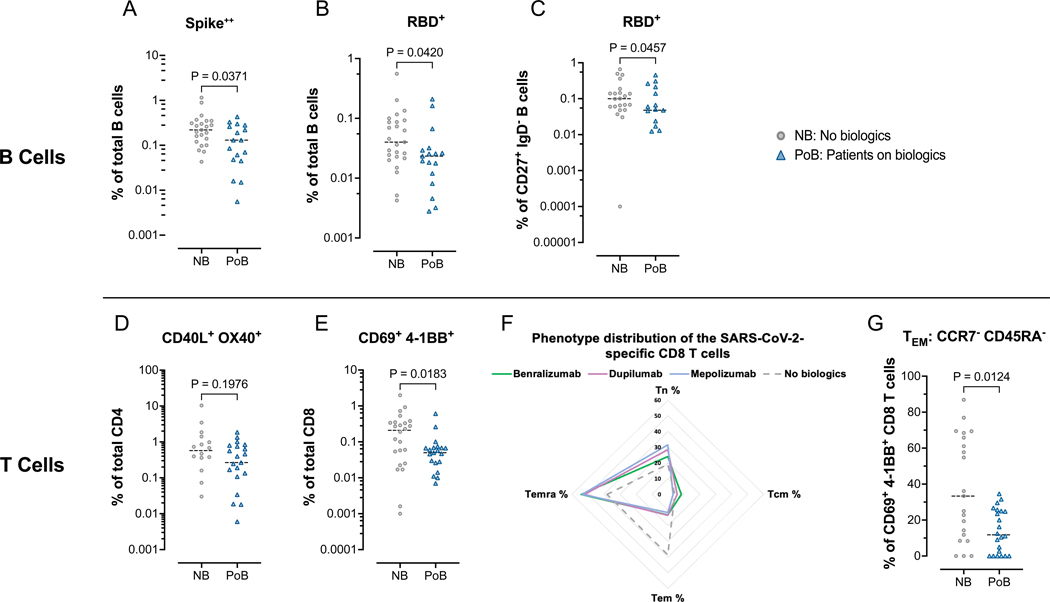
Figure 6. Flow cytometry assessment of SARS-CoV-2-specific B and T lymphocytes.
All data points in this figure are from days 130–250 after V2 (A-C) Fluorescently-conjugated Spike and RBD proteins in a tetramer conformation were used to identify ex-vivo SARS-CoV-2-specific B cells. All Spike-specific cell populations were identified using double-labeling of fluorochromes. (A) Percentages of Spike positive cells of total B cells. (B) Percentages of RBD positive cells of total B cells. (C) Percentages of RBD positive cells of switched memory B cells (CD27+, IgD–). (D-G) Ex-vivo PBMCs from NB and PoB were incubated for 24-hours with Spike peptides, with Staphylococcal enterotoxin B as a positive control, or with no antigens as a negative control. SARS-CoV-2 specific T cells were quantified using markers of activation (CD40L and OX40 for CD4 T cells, and CD69 and 4–1BB for CD8 T cells). SARS-CoV-2 specific values are calculated and plotted as the experimental condition with Spike peptides minus the experimental condition without peptides (negative control). (D) Percentages of CD40L+ OX40+ cells of total CD4 T cells. (E) Percentages of CD69+ 4–1BB+ cells of total CD8 T cells. (F) Radar plot showing the phenotypic distribution of SARS-CoV-2-specific CD8 T cells. (G) Comparisons of TEM percentages of the SARS-CoV-2 specific CD8 T cells. PBMC, peripheral blood mononuclear cells; NB, control subjects not on biologics; PoB, patients on biologics include all subjects treated with benralizumab, dupilumab and mepolizumab; V2, dose two of the SARS-CoV-2 mRNA vaccine; WT, wild-type; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RBD, receptor-binding domain; mRNA, messenger ribonucleic acid; TEM, effector memory CCR7– CD45RA–; TCM, central memory CCR7+ CD45RA–; TEMRA, terminally differentiated effector CCR7– CD45RA+, TN, naïve CCR7+ CD45RA+.