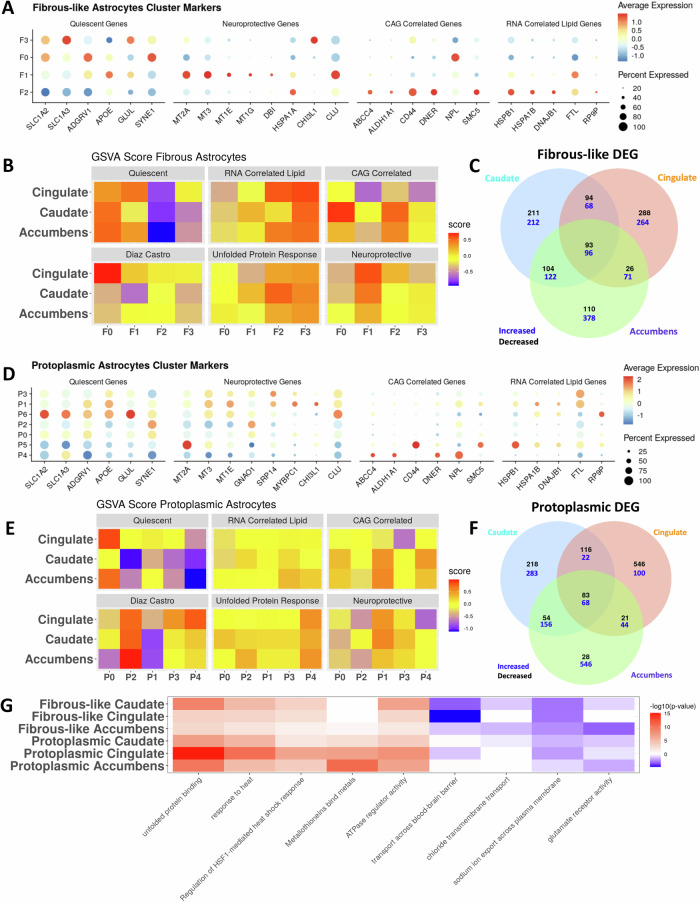
Fig. 4. Astrocytes are regionally heterogeneous in HD.
A Dot plot displaying the expression of select genes in fibrous-like astrocytic clusters. The genes were selected from four gene sets (Quiescent: baseline astrocyte genes, Neuroprotective: predicted from our previous work – see main text, CAG-Correlated: genes with significant positive regression weights - see Fig. 1E for more details, RNA-Correlated Lipid.: set of genes that correlated with lipid abundance from Fig. S3D. B Heatmap of the average enrichment scores determined by gene set variation analysis of select gene sets within each fibrous-like astrocytic sub-cluster per brain region. The gene sets of interest include the four in A, a core astrocytic signature described in Diaz-Castro et al. 20, and a GO term for response to unfolded protein (GO:0006986). C Venn diagram showing overlap between the differentially expressed genes (DEGs) in HD vs control in all fibrous-astrocytes across the three brain regions (increased: blue; decreased: black). D Similar to (A) but for protoplasmic astrocytic sub-clusters. E Similar to (B) but for protoplasmic astrocytic sub-clusters. F Similar to (C) but for protoplasmic astrocytes. G Heatmap displaying the negative log10(p-value) of enrichment of select GO terms (columns) in DEGs from fibrous-like and protoplasmic astrocytes DEGs per region (C, F) – rows. Red indicates terms significantly enriched in DEGs increased in HD, blue indicates GO terms enriched in DEGs significantly decreased in HD, and white indicates no significance. P value adjustment was done in gprofiler using the g:SCS method.