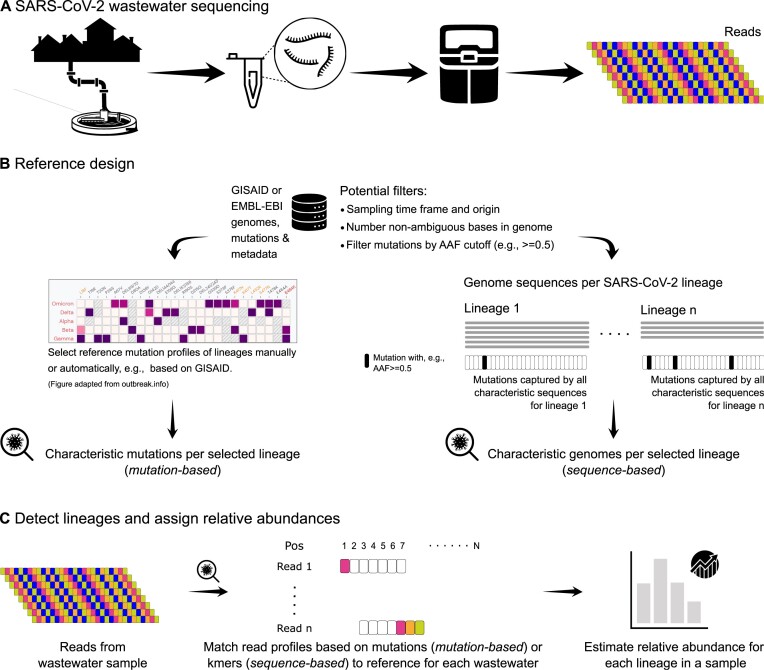
Figure 1:
Schematic overview of reference design and lineage abundance estimation from SARS-CoV-2 wastewater sequencing data. (A) Wastewater samples are collected from sewer influent, for example. RNA is extracted and, in the context of SARS-CoV-2, usually amplified as cDNA using established primer schemes and then sequenced to obtain short snippets of viral RNA (reads). (B) Current methods (Table 1 for lineage assignment and abundance estimation) need a reference dataset, usually constructed from genomes and mutations derived from clinical sequencing and patient samples. Here, we distinguish 2 general approaches to design the reference, where either marker mutations are preselected (mutation based) or full-genome sequences are selected (sequence based). (C) The data analysis part may differ considerably depending on the implementation. However, all tools attempt to assign known lineages and estimate their frequency in the mixed sample based on mutations that can be detected in the reads. Our study uses MAMUSS as an exemplary mutation-based approach based on a 2-indicator classification and preselected marker mutations characteristic for certain lineages [20]. For the sequence-based approach, we use a Nextflow implementation (VLQ-nf) of the slightly adjusted VLQ pipeline as proposed by Baaijens et al. [29] and is based on the tool Kallisto. AAF: alternative allele frequency, used as a cutoff to define a mutation as a feature.