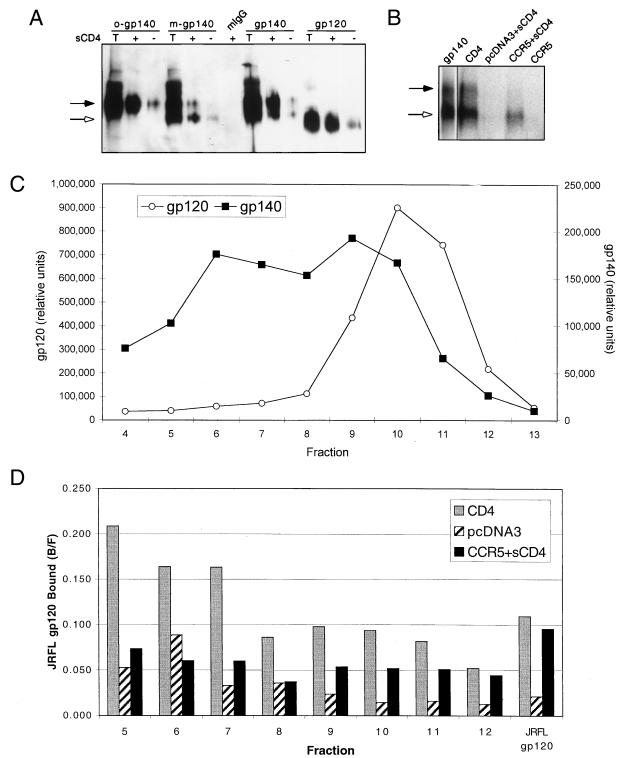
FIG. 7.
(A) Reactivity of JRFL gp140 with MAb 17b. Purified JRFL gp140 protein was fractionated by sucrose gradient sedimentation. Representative fractions containing exclusively oligomeric (o-gp140) or exclusively monomeric (m-gp140) gp140 were immunoprecipitated with the MAb 17b or with mouse IgG (mIgG) in either the presence (+) or the absence (−) of sCD4. Unfractionated gp140 and gp120 samples were also immunoprecipitated. Briefly, 50 μl of sucrose gradient fractions or 100 ng of purified protein was combined with 500 ng of sCD4 (where indicated). After 30 min at room temperature, 20 μl of ProG beads, 1 μg of MAb, 20 μl of 5% BSA, and PBS were added for a total volume of 400 μl, rocked at RT for 2 h, and washed twice with PBS. The samples were run on an 8% SDS-polyacrylamide gel along with a sample of the total input protein (T) and probed with an anti-gp120 rabbit sera by Western analysis. We note that gp120 produced as a gp140 precursor migrated slower than gp120 produced alone, most likely due to differences in glycosylation. (B) Binding of gp140 to CCR5. Radiolabeled JRFL gp140 was bound to 5 × 105 cells expressing the indicated receptors for 2 h as described in Materials and Methods. Instead of being washed through glass fiber filters, however, cells were washed twice with 1 ml of wash buffer in Eppendorf tubes. The protein bound to the cells was detected by lysing the cells in 25 μl of 0.5% Triton X-100, spinning out the cell debris, loading the supernatant onto an 8% SDS-polyacrylamide gel, and visualizing the protein by autoradiography. Approximately 25% of gp140 is binding to CCRS compared to CD4, as measured by PhosphorImager analysis and adjusting for total Env. The gp140 lane is a separate exposure of the initial protein added. The black arrows in panels A and B indicate the location of gp140, and the white arrows indicate the location of gp120. (C) Radiolabeled JRFL gp140 was separated through a 5 to 20% continuous sucrose gradient as previously described (13). A sample of each fraction was loaded onto an SDS-polyacrylamide gel, separated, and visualized by autoradiography. The amount of gp120 and gp140 in each fraction was quantitated by PhosphorImager densitometry. (D) A sample of each fraction from the gp140 sucrose gradient in panel C was bound to cells expressing CD4, CCR5, or pcDNA3 as indicated. The fraction bound (bound/free [B/F]) is indicated for each sample. The results from early fractions appear relatively high in value due to the lower counts of radioactivity in these fractions. All results should therefore be evaluated relative to background (pcDNA3) binding. All results are from representative experiments repeated twice.