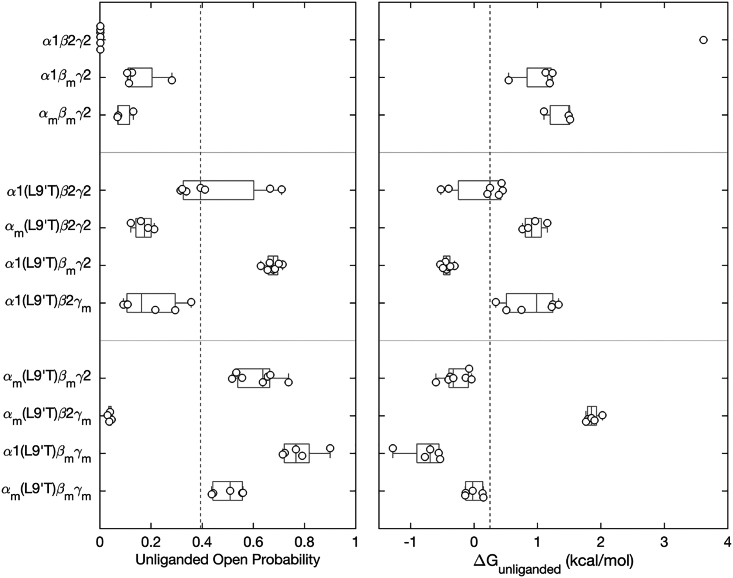
Figure 6.
M2-M3 linker mutations either enhance (βm) or inhibit (αm, γm) intrinsic pore opening in the absence of ligand. Left: spontaneous unliganded open probability for receptors with M2-M3 linker mutations αm, βm, and/or γ m in the gain-of-function α1(L9′T)β2γ2 or wild-type α1β2γ2 backgrounds estimated as the ratio of PTX-sensitive to maximal GABA-evoked current amplitudes (see Figs. 2 and 4). Right: free energy difference from closed to open states calculated from the unliganded open probability using Eq. 2 (see materials and methods). for α1β2γ2 receptors is based on the estimate (46), which is undetectable in our assay. All other data points are for individual oocytes (α1β2γ2: n = 6; α1βmγ2: n = 4; αmβmγ2: n = 3; α1(L9′T)β2γ2: n = 7; αm(L9′T)β2γ2: n = 4; α1(L9′T)βmγ2: n = 7; α1(L9′T)β2γm: n = 6; αm(L9′T)βmγ2: n = 7; αm(L9′T)β2γm: n = 5; α1(L9′T)βmγm: n = 5; αm(L9′T)βmγm: n = 5). Boxplots indicate quartiles, and the vertical dashed line is the median for the α1(L9′T)β2γ2 background. The unliganded open probabilities for some of the constructs are the same as shown in Figs. 3 and 4.