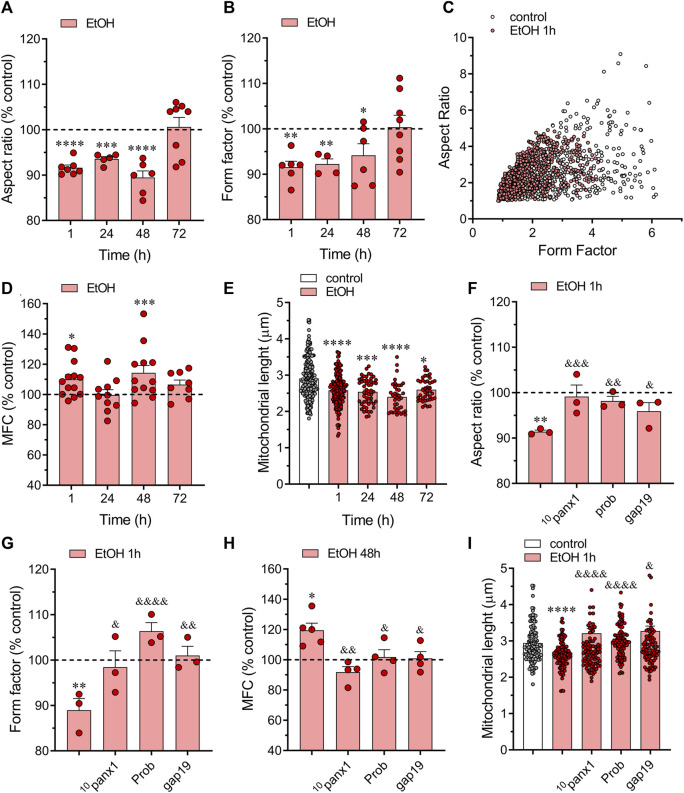
FIGURE 5.
Panx1 and Cx43 hemichannels contribute to the ethanol-induced mitochondrial fragmentation in astrocytes. (A, B) Averaged data normalized to control conditions (dashed line) of aspect ratio (A) and form factor (B) by astrocytes treated with 100 mM ethanol for several time periods. (C) Scatter plot of form indexes distribution of individual mitochondria from a representative control (white circles) and 100 mM ethanol-treated astrocyte for 1 h (red circles). (D) Averaged data normalized to control conditions (dashed line) of mitochondrial fragmentation count (MFC) by astrocytes treated with 100 mM ethanol for several time periods. (E) Averaged data of mitochondrial length by astrocytes under control conditions (white violin) or treated with 100 mM ethanol for several time periods (red violins). Each dot represents the cell-averaged mitochondrial length of 20 mitochondria per cell. Overall, these dots encompass at least five independent culture experiments from different litters of animals. (F–H) Averaged data normalized to control conditions (dashed line) of aspect ratio (F), form factor (G), and MFC (H) by astrocytes treated for 48 h with 100 mM ethanol alone or plus the following pharmacological agents: 100 µM 10panx1, 500 µM probenecid (Prob) or 100 µM gap19. (I) Averaged data of mitochondrial length by astrocytes under control conditions (white violin) or treated for 48 h with 100 mM ethanol alone (red violin) or plus the following pharmacological agents: 100 µM 10panx1, 500 µM probenecid (Prob) or 100 µM gap19. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ethanol compared to control conditions; & p < 0.05, && p < 0.01; &&& p < 0.001, &&&& p < 0.0001; effect of pharmacological agents compared to ethanol treatment (one-way ANOVA followed by Tukey’s post hoc test). Each dot represents an independent culture experiment from a different litter of animals. The value reflects the mean of at least three coverslip replicates, with a minimum of 60 cells analyzed per coverslip.