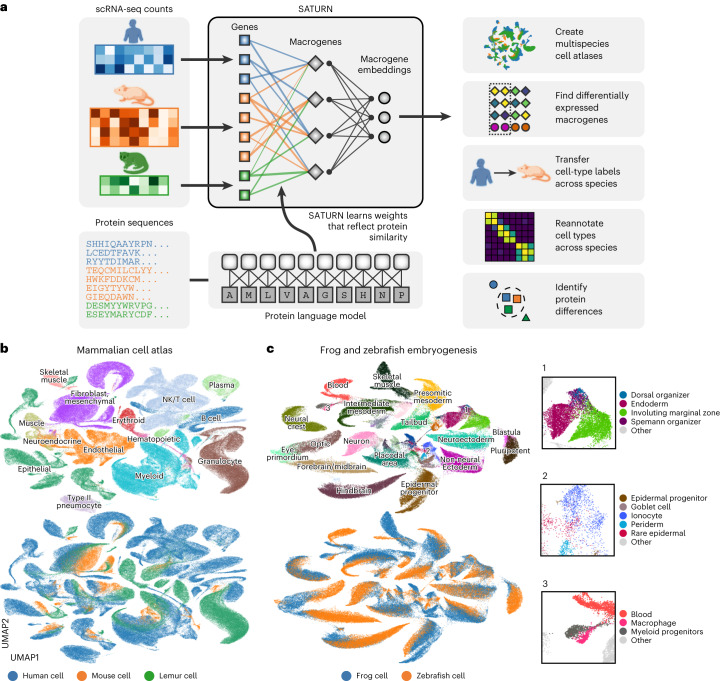
Fig. 1. SATURN incorporates protein sequences and gene expression to embed single cells.
a, Overview of SATURN. SATURN takes as input scRNA-seq datasets generated from one or more species and the amino acid sequences of proteins present in these species. SATURN then maps each species’ genes to a joint feature space by learning ‘macrogenes’, that is, groups of functionally related intraspecies and interspecies genes. Finally, in the shared macrogene space, SATURN integrates datasets across species by learning a joint cell embedding space in which cell types conserved across species are aligned with each other. b, UMAP visualization of a joint embedding space across three distinct species. We applied SATURN to integrate cell atlas datasets of 335,000 cells from Tabula Sapiens (human), Tabula Microcebus (mouse lemur) and Tabula Muris (mouse), creating a mammalian cell atlas. Colors denote coarse-grained cell-type annotations (top) and species annotations (bottom). Only cell types with more than 350 cells were included. c, UMAP visualization of SATURN’s integration of datasets from frog (97,000 cells) and zebrafish (63,000 cells) embryogenesis. Colors denote different major cell types (top) and different species (bottom). In SATURN’s embedding space, cell types conserved across species aligned well (for example, frog/zebrafish neural crest), while species-specific cell types formed separate single-species clusters (for example, frog goblet cells). Cell types not directly mapped between both species shared similar ontology, for example, the zebrafish dorsal organizer and frog Spemann organizer (inset 1). Epidermal cell types including periderm, epidermal progenitor and rare epidermal cell types were also aligned, as were specialized epithelial cells such as goblet cells and ionocytes (inset 2). Finally, myeloid cell types including macrophages and myeloid progenitors clustered together (inset 3).