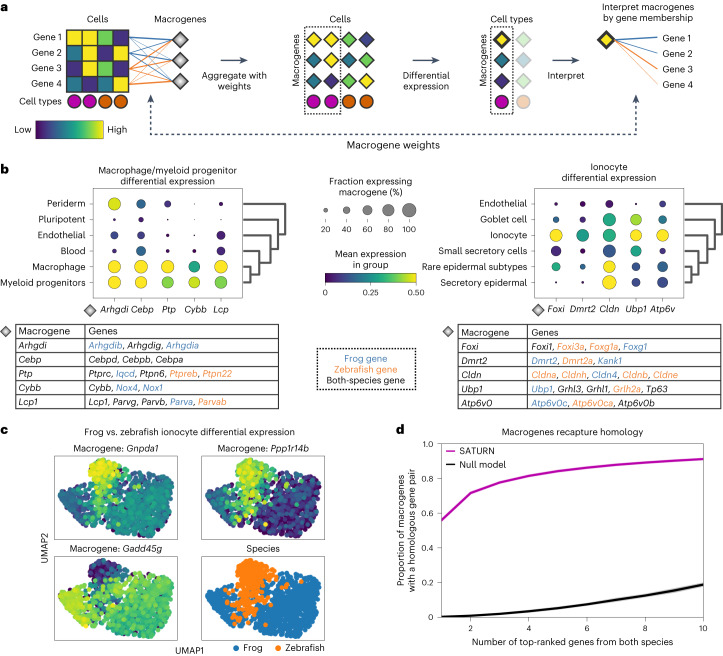
Fig. 2. SATURN enables multispecies differential expression analysis in the macrogene space.
a, Overview of SATURN’s differential expression analysis on macrogenes. Every gene is connected to a macrogene with a corresponding weight that represents the importance of that gene to the given macrogene. Thus, each cell has corresponding macrogene values calculated as the weighted and normalized sum of its gene expression values. Because SATURN operates in the macrogene space, differential expression for resulting cell clusters gives the set of differentially expressed macrogenes of a given cell type. Finally, the genes with the highest weights to a macrogene are used to interpret the macrogene. b, Differentially expressed macrogenes on frog and zebrafish embryogenesis datasets for macrophage and myeloid progenitors (left) and ionocytes (right). Differential expression is performed by comparing these cell types with all other cell types. We show only cell types that are similar to target cell types determined as expressing a subset of the top differentially expressed macrogenes. We assigned names to macrogenes based on the set of genes with the highest weight in the given macrogene. The tables show the top five differentially expressed macrogenes and the top weighted genes in each macrogene. Genes are shown in black if a gene is included in the top genes for both species in a given macrogene, and blue or orange if the gene is frog or zebrafish specific, respectively. c, Macrogene differential expression can also be used to find species-level differences between cell types conserved across species. Example of differentially expressed macrogenes between frog and zebrafish ionocytes. d, SATURN macrogenes contained a far higher proportion of homolog gene pairs than what would be expected by chance, demonstrating that SATURN recaptures sequence-based homology. The purple curve shows the proportion of SATURN macrogenes that contain, within their top-ranked frog and top-ranked zebrafish genes, at least one homolog gene pair, versus the top number of genes. Homology was determined according to BLASTP results. The black curve shows the proportion obtained by a null model in which the same number of genes are randomly selected without replacement from both species.