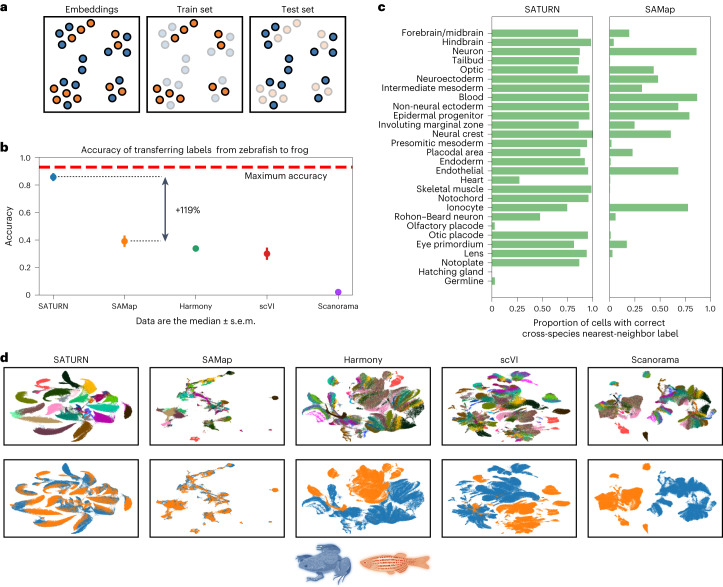
Fig. 3. SATURN embeddings capture shared cell-type identity in frog and zebrafish embryogenesis.
a, Explanation of how multispecies embeddings are scored. A joint embedding space, containing cells from multiple species, is split by species into a training set and a test set. A classification model to predict cell types is trained on a single-species training set, and evaluated on the test set of another species. The maximum test set accuracy achievable will be lower than 100% if the test set species contains specific cell types that cannot be predicted by a classifier trained on the training species. Blue denotes frog, while orange denotes zebrafish. b, Median performance of SATURN compared to alternative methods. The performance is evaluated using the prediction accuracy of a logistic classifier model trained to differentiate zebrafish cell types and tested on predicting the cell-type annotations of frog cells. Higher values indicate better performance, and 0.93 is the maximum accuracy that can be reached by label transfer on this dataset. SAMap represents a version of the SAMap method in which cell-type annotations are used to integrate datasets. Vertical position of scatterplot points represents the median accuracy score across 30 runs for each method. Error bars represent standard error. For batch correction methods (Harmony, scVI and Scanorama), the input genes are selected as the one-to-one homologs determined by ENSEMBL. c, SATURN produces more homogeneous clusters than SAMap, and these clusters contain accurate multispecies cell types. Bars represent the percentage of cells from zebrafish that are nearest neighbors of frog cells of the given cell type conserved across these two species. Cell types are ordered by frequency. d, Comparison of UMAP visualizations of integrated frog and zebrafish embryogenesis datasets generated by SATURN and alternative methods. In SATURN’s embedding space, different cell types naturally form clusters and cells from different species align well. On the other hand, alternative baselines either do not preserve cell-type information (SAMap) or cannot integrate two species (Harmony, scVI and Scanorama).