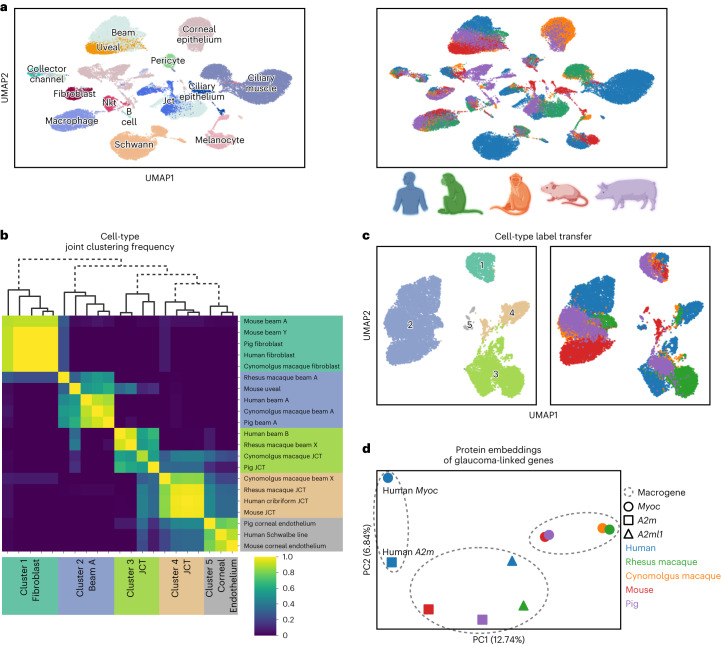
Fig. 4. SATURN discovers new cell types and facilitates the analysis of protein embeddings for the AH cell atlas.
a, SATURN successfully aligned 50,000 cells from the AH cell atlas consisting of five species: human, cynomolgus macaque, rhesus macaque, mouse and pig. UMAP visualization of SATURN’s embeddings where colors denote cell types (left) and species (right). b,c, We applied SATURN to regroup cell types in a multispecies context. By clustering SATURN’s embeddings, we found five broad cell types. b, Heat map and dendrogram of reannotated cell types using SATURN. Labels on the right side show original cell-type annotations, while on the bottom we show reannotations obtained using SATURN. These clusters include cell types originally labeled as fibroblast and beam A/Y cells (cluster 1), beam A and uveal cells (cluster 2), JCT and beam cells (cluster 3 and cluster 4) and corneal endothelium cells (cluster 5). Across 30 independent experiments, we regrouped cluster 1 as fibroblast cells, cluster 2 as beam A cells, clusters 3 and 4 as JCT cells, and cluster 5 as corneal endothelium cells. We specifically reannotated mouse beam A and beam Y cells, which have high expression of fibroblast markers such as Pi16, Fbn1 and Mfap5 as originally noted18. We additionally regrouped human beam B cells, which were not found in other species, as JCT cells. Finally, we mapped beam X cells, which were unique to rhesus and cynomolgus macaque, to two JCT clusters. c, UMAP visualizations of reannotated cell types. Cells are colored according to annotations inferred by SATURN (left) and species information (right). d, SATURN facilitates the analysis of protein embeddings by creation of multispecies macrogenes. Human MYOC had the highest weight to a different macrogene than the other four species’ Myoc variants. The human gene A2M also had the highest weight to the human MYOC macrogene. We can investigate this discrepancy by visualizing the protein embeddings of Myoc and A2m from all five species using principal component analysis. This analysis offers potential to point to similar function in A2m as Myoc, which would otherwise not be identified by sequence-based homology, as well as potential differences in human MYOC and Myoc in the other four species.