FIGURE 2.
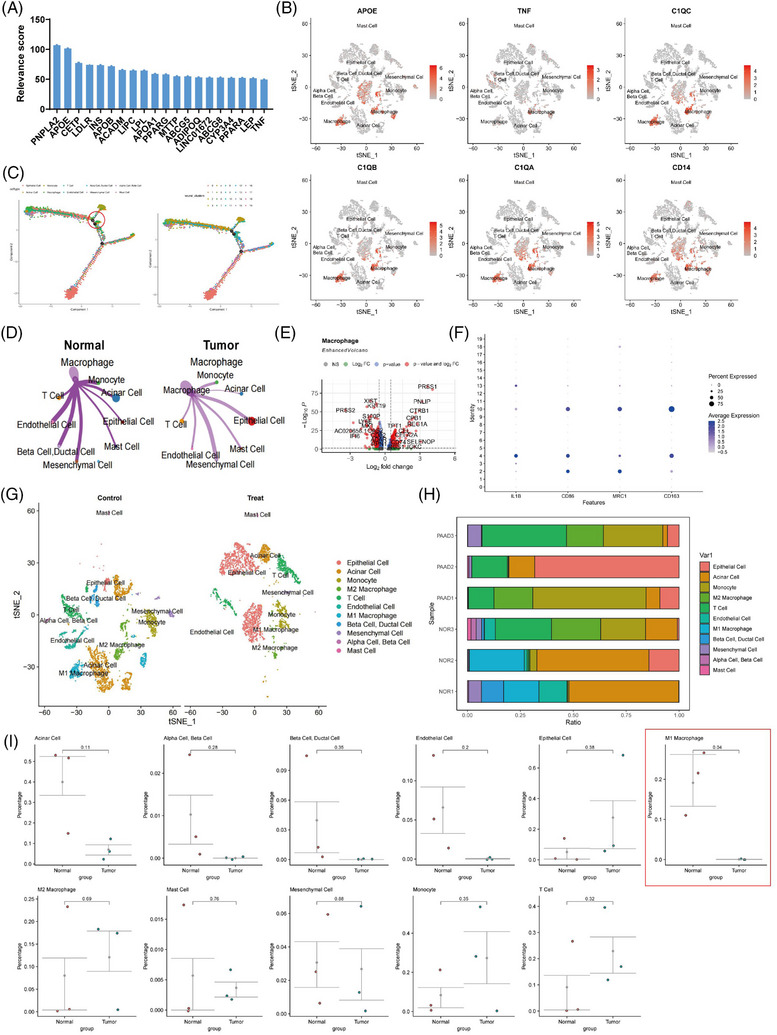
Study on the differential molecular characterisation of macrophages in pancreatic cancer tissues. (A) The relevance scores of the top 20 genes in the ‘LIPID METABOLIC PROCESS’ gene set in the Genecards database. (B) Expression profiles of macrophage marker genes in different cell subpopulations in adjacent normal tissues and pancreatic adenocarcinoma (PAAD) tissues samples (N = 6), with darker red indicating higher average expression levels. (C) Construction of cell trajectories based on cell types and clustering (N = 6). (D) Differential cell‒cell communication map of macrophages in adjacent normal tissue samples compared to PAAD tissue samples, where line thickness represents interaction strength (N = 6). (E) Volcano plot of differentially expressed genes between macrophages in adjacent normal tissue samples and PAAD samples, with red dots on the left of the dashed line indicating genes with higher expression in PAAD samples and dots on the right indicating genes with lower expression in PAAD samples (N = 6). (F) Expression profiles of known M1 macrophage‐specific marker genes in different clusters in adjacent normal tissue (normal, N = 3) and PAAD samples (tumour, N = 3), with darker blue indicating higher average expression levels and larger circles representing more cells expressing that gene. (G) Visualisation of cell annotations based on t‐SNE clustering for normal group (N = 3) and PAAD group (N = 3), with each colour representing a cell subpopulation. (H) Proportion of different cell subpopulations in each sample, represented by different colours (N = 6). (I) Differences in cell composition between adjacent normal tissue and PAAD samples analysed by t‐test, with control (N = 3) representing adjacent normal tissue samples, treat (N = 3) representing PAAD tissue samples, and p‐values indicating the significance level of differences in cell composition between control and treat groups.
