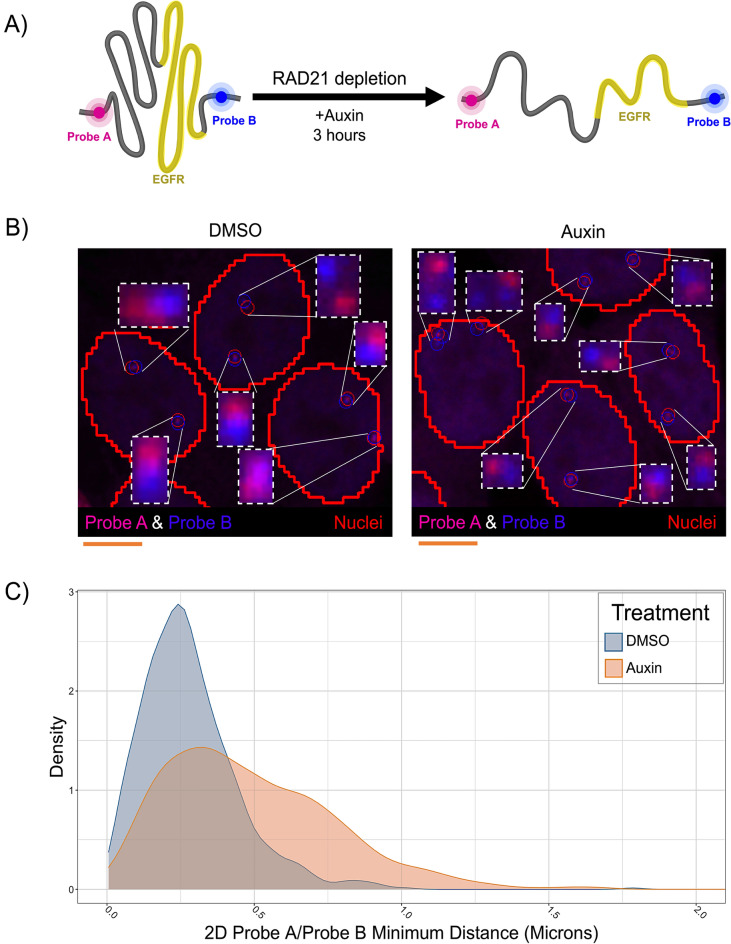
Figure 3.
HiTIPS measures distances between genomic loci labeled with High-Throughput DNA FISH probes. (A) Schematic representation of the experiment to measure spatial distances of two genomic loci located at the base of a TAD encompassing the EGFR gene on Chr 7 and detected by FISH probes in different channels (Probe A and Probe B). Treatment with Auxin in HCT116-RAD21-AID cells leads to rapid proteolytic degradation of the RAD21-AID fusion protein via the ubiquitin/proteasome pathway. (B) Representative 3D maximally projected images of HCT116-RAD21-AID cells stained with DNA FISH probes A and B targeted to the 5’ and 3’ boundaries of the EGFR TAD, respectively, and of the results of the HiTIPS spot finding algorithm results overlaid as red and blue circles, respectively. Scale bar: 5 microns. (C) Density plots of minimum spatial distances between the A and B DNA FISH probes in 1874 cells treated with Auxin or DMSO.