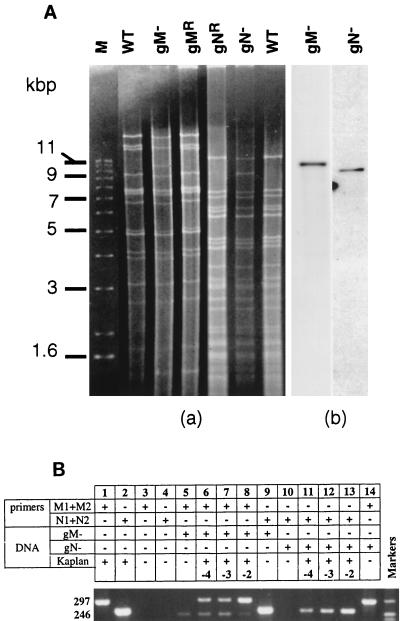
FIG. 2.
Genotype assessment of the viral stocks. (A) Southern analysis of wild-type (WT) PrV, gM− and gN− mutants, and revertants. After extraction, viral DNA was digested with either BamHI (WT, gM−, and gMR) or PstI (gNR, gN−, and WT). Fragments were separated on a 0.6% agarose gel containing ethidium bromide (gel a), transferred to a nylon membrane, and hybridized with a LacZ probe (pGEM 3Zf; Promega) labeled with digoxigenin (gel b). Size markers (M) are on the left. (B) PCR amplifications were done with around 20 ng of wild-type or mutant DNA, 1 pmol of each primer, 2.5 U of Taq DNA polymerase (Appligene), 50 μM concentration of each deoxynucleoside triphosphate, and 10% dimethyl sulfoxide per reaction mixture. In addition to mutant DNA, 2, 20, and 200 pg of wild-type PrV DNA were added to samples 6 and 11, 7 and 12, and 8 and 13, respectively. Thirty cycles of amplification at 95, 50, and 72°C for 60, 40, and 120 s, respectively, were performed. PCR products were separated in 2% agarose gels containing ethidium bromide and were visualized in UV light. A pBR322 DNA-MspI digest was used for size markers (sizes of the bands, 217, 238, 242, and 307 nucleotides). Numbers at the left are molecular sizes of the amplified products, in nucleotides.