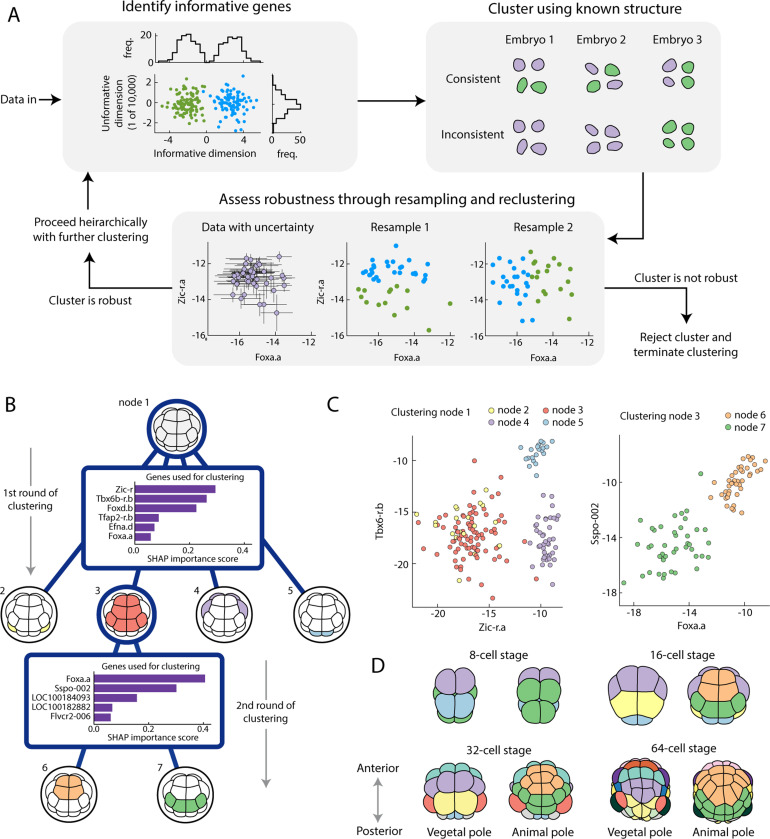
FIG. 2. Principled and physically motivated clustering robustly recovers cell types.
(A) Overview of clustering algorithm highlighting the three key underlying principles. 1. Identify informative genes. Informative dimensions split the data into separable clusters (green vs blue points, top left), uninformative dimensions do not, and make identifying clusters harder. 2. Cluster using known structural priors. A cell type should have equal numbers of cells in each embryo, a clustering without that property is inconsistent (purple and green cells represent clusters, top right). 3. Assess robustness through resampling and reclustering. We have a posterior for our expression levels [23] (purple points with error bars, bottom), and by repeatedly resampling and clustering (blue vs green points, bottom), we assess how consistent the clusters are and decide whether to keep or reject a cluster. For full details see SM Sec. III. (B) Worked example of the hierarchical clustering for the 16-cell stage. Starting from all cells at node 1, the algorithm splits into 4 clusters using the 6 genes shown. The SHAP importance score is an estimated measure of how useful each gene is when performing this round of clustering [30] (SM Sec. III). The algorithm then splits cluster 3 into two further clusters using a different set of 5 genes. The algorithm terminates here as further clusters are assessed to be not robust (SM Sec. III). (C) The two most important genes shown for both clustering stages, showing separation between identified clusters (node 2 and 3 differ in their expression of Foxd.b). (D) 8-cell through 64-cell stages colored by cell type, showing increasing transcriptomic specialization in time.