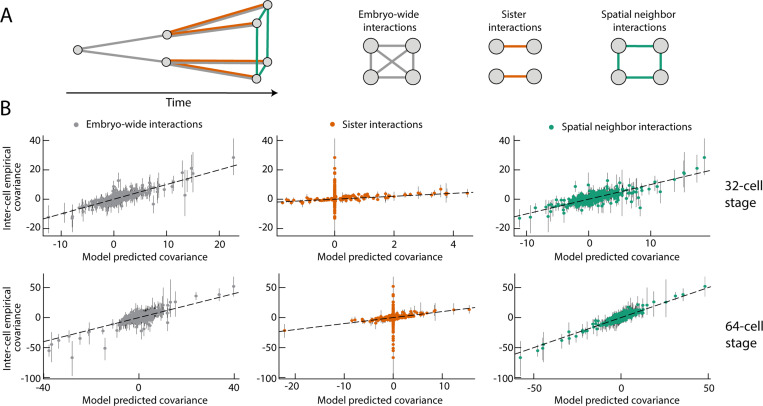
FIG. 6. Comparing models with data identifies plausible families of statistical physical models for interactions.
(A) Correlations between different cells could arise from signaling resulting in interactions between spatial neighbors, cell division resulting in interactions between sister cells, or variations in the zygote resulting in all-to-all coupling across the embryo. We consider how well these different hypotheses explain the experimental data. (B) The inter-cell empirical covariances (IEC) are covariances between different cell types measured from data. The model predicted covariances (MPC), are covariances between different cell types in the maximum entropy model (Fig. 5D). A model which explains the data should have MPC values equal to the IEC values. Here, we test models which assume that all cells within an embryo directly interact (gray) only sisters interact (orange), or only spatial neighbors interact (green). Both the embryo-wide and spatial model are able to reasonably explain the data, with points lying around (black dashed line), whereas sister-cell interactions alone are not sufficient. Without interactions between non-sister cells, many covariances are zero in the model despite being non-zero in the data. Error bars are from the 25th to 75th percentile of covariance estimates.