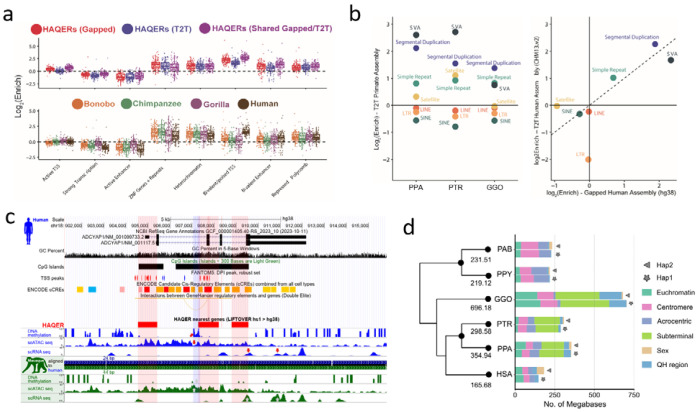
Figure 5. Divergent regions of the ape genomes.
a) HAQER (human ancestor quickly evolved region) sets identified in gapped (GRCh38) and T2T assemblies show enrichments for bivalent gene regulatory elements across 127 cell types and tissues, with the strongest enrichment observed in the set of HAQERs shared between the two analyses (top). The tendency for HAQERs to occur in bivalent regulatory elements (defined using human cells and tissues) is not present in the sets of bonobo, chimpanzee, or gorilla AQERs (ancestor quickly evolved regions; bottom). b) AQERs are enriched in SVAs, simple repeats, and SDs, but not across the general classes of SINEs, LINEs, and LTRs (left). With T2T genomes, the set of HAQERs defined using gapped genome assemblies became even more enriched for simple repeats and SDs (right).c) HAQERs in a vocal learning-associated gene, ADCYAP1 (adenylate cyclase activating polypeptide 1), are marked as containing alternative promoters (TSS peaks of the FANTOM5 CAGE analysis), candidate cis-regulatory elements (ENCODE), and enhancers (ATAC-Seq peaks). For the latter, humans have a unique methylated region in layer 5 extra-telencephalic neurons of the primary motor cortex. Tracks are modified from the UCSC Genome Browser75 above the HAQER annotations and the comparative epigenome browser76 below the HAQER annotations. d) Lineage-specific structurally divergent regions (SDRs). SDRs are detected on two haplotypes and classified by different genomic content. The average number of total bases was assigned to the phylogenetic tree.