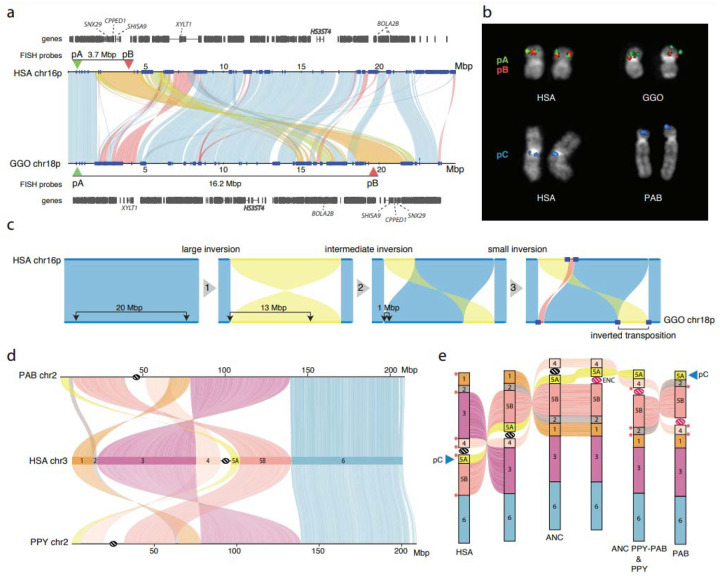
Figure 4. Great ape inversions and evolutionary rearrangements.
a) Alignment plot of gorilla chr18p and human chr16p shows a 4.8 Mbp inverted transposition (yellow). SDs are shown with blue rectangles. b) Experimental validation of the gorilla chr18 inverted transposition using FISH with probes pA (CH276–36H14) and pB (CH276–520C10), which are overlapping in human metaphase chromosomes. The transposition moves the red pB probe further away from the green pA probe in gorilla, resulting in two distinct signals. FISH on metaphase chromosomes using probe pC (RP11–481M14) confirms the location of a novel inversion to the p-ter of PAB chr2. c) An evolutionary model for the generation of the inverted transposition by a series of inversions mediated by SDs. d) Alignment plot of orangutan chromosome 2 homologs to human chromosome 3 highlights a more complex organization than previously known by cytogenetics45: a novel inversion of block 5A is mapping at the p-ter of both chr2 in PAB and PPY. e) A model of serial inversions requires three inversions and one centromere repositioning event (evolutionary neocentromere; ENC) to create PPY chromosome 2, and four inversions and one ENC for PAB. Red asterisks show the location of SDs mapping at the seven out of eight inversion breakpoints.