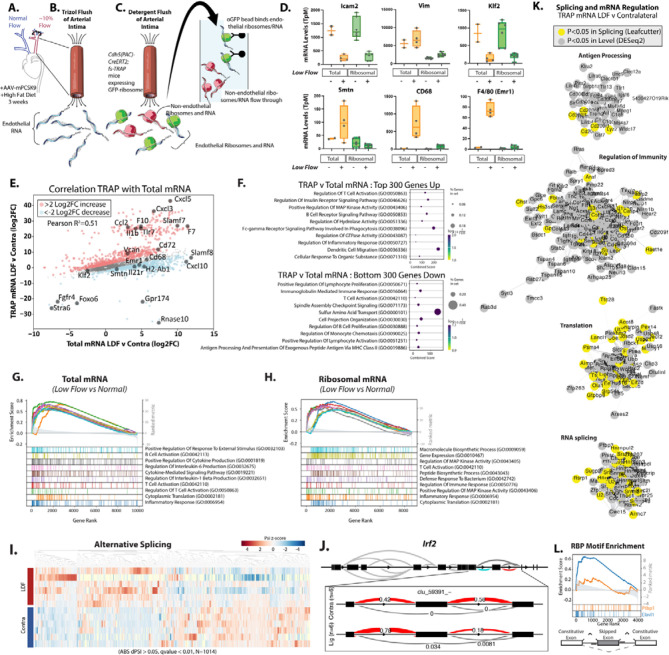
Figure 1. Translating ribosome affinity purification (TRAP) from atherogenic endothelium identifies changes in immune-regulatory pathways.
(A) Partial Carotid Arterial Ligation (PCAL) induction of low flow in the left carotid artery (LCA) and hyperlipidemia collected at 3 weeks post PCAL. (B) Approach for isolating total intimal mRNA from both low and disturbed flow (LDF) and contralateral normal flow vessels via Trizol flush. (C) Approach for isolating endothelial-specific ribosomal mRNA from both low flow and normal flow vessels via RiboTRAP. (D) Expression level (FPKM) of indicated genes in low flow and normal flow vessels. (E) Plot showing the effects on transcript levels between LDF and contralateral arteries, between TRAP (Y-axis) and total mRNA (X-axis). (E) mRNA expression (Transcript per Million) detected in Total intimal RNAseq compared to TRAP RNAseq. (F-H) Gene Set Enrichment Analysis (GSEA) of low flow regulated transcripts thresholded on mean TpM greater than or equal to 100. Top 300 genes up indicate the genes and associated transcripts most enriched in TRAP low vs. contralateral relative to Total mRNA low vs. contralateral (red in E), down is depleted in TRAP vs. contralateral (blue in E). (G&H) GSEA plot of top 10 upregulated gene sets in GoTerms in (G) total mRNA and (H) ribosomal mRNA. (I&J) Leafcutter analysis of alternative splicing in ribosomal associated mRNA, showing clustering of groups by alternative splicing levels (I), where dPsi indicated the change in inclusion frequency between low flow and contralateral artery, plotted here as Z-scores and an example plotted splicing event (J). (K) Cytoscape visualization of top categories associated with low flow regulated transcripts in ribosomal mRNA. (L) Enrichment of Cis-RNAbp motifs nearby regulated skipped exons, showing the relative enrichment for Elavl1 and Ptbp1 6mers. (A-L) Total intimal RNAseq (Total; N=4 WT Ligated vs. N=2 Contralateral) compared to TRAP RNAseq (TRAP; N=6 WT Ligated vs. N=6 Contralateral) (K) Grey = differential mRNA TRAP, DESeq2 p.adjusted <0.05, yellow = differential mRNA splicing Leafcutter p.adjusted <0.05.