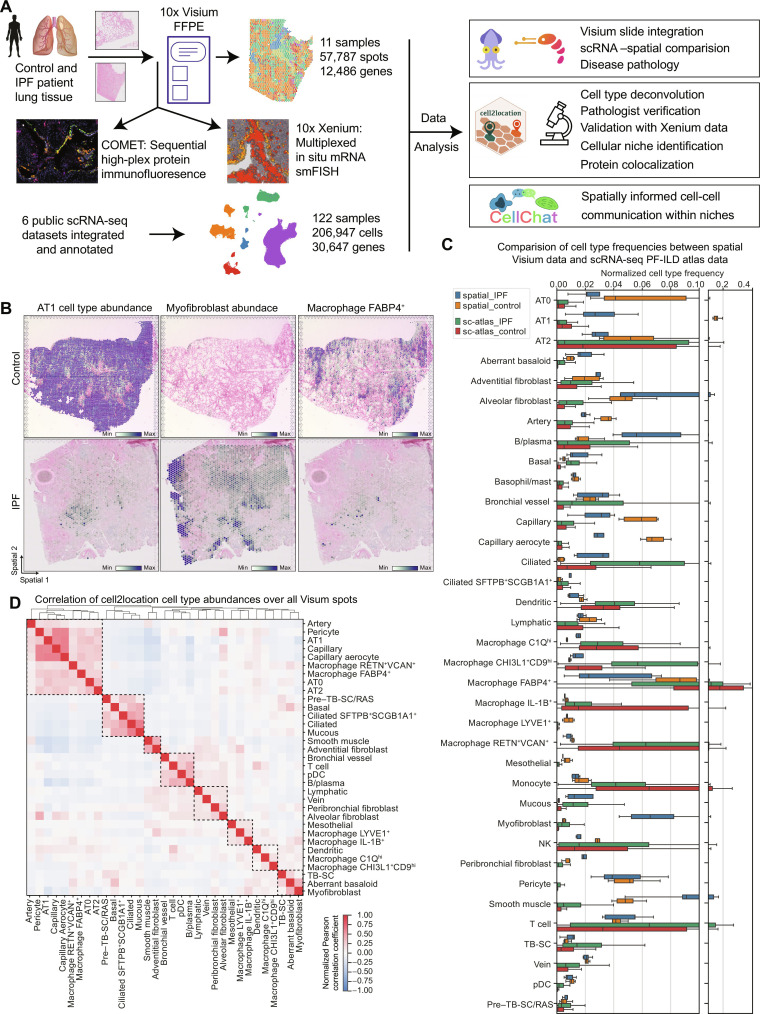
Fig. 1. Combining spatial transcriptomics and scRNA-seq places genes and cell types to their tissue location.
(A) Experimental design included spatial transcriptomics using Visium for FFPE and CytAssist (10x Genomics) on control and IPF patient samples (seven donors, 4× sections Visium CytAssist for FFPE, and 7× sections with Visium for FFPE; see fig. S1A for distribution of donors and sections). On two adjacent IPF and two control sections, as well as three additional IPF sections, we applied Xenium in situ (10x Genomics) for multiplexed in situ mRNA imaging similar to Single Molecule Fluorescence In Situ Hybridization (smFISH) and high-plex sequential protein immunofluorescence on the COMET platform. Six publicly available scRNA-seq datasets were integrated and annotated into a PF-ILD atlas. Visium and scRNA-seq data were combined for various analysis steps, including cell type deconvolution or cell-cell communication. (B) Spatial plots show cell2location mapping of estimated cell type abundance of AT1, FABP4+ alveolar macrophages, and myofibroblast cells on one control and one IPF tissue section. (C) Comparison of normalized cell type frequencies in spatial transcriptomic data and scRNA-seq PF-ILD atlas. (D) Pearson correlation of cell type abundances estimated with cell2location across all tissue sections identifies spatial colocated cell type modules.